+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1p53 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Crystal Structure of ICAM-1 D3-D5 fragment | |||||||||
 Components Components | Intercellular adhesion molecule-1 | |||||||||
 Keywords Keywords |  CELL ADHESION / CELL ADHESION /  IgSF domain / IgSF domain /  beta-sheet / dimer beta-sheet / dimer | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of leukocyte mediated cytotoxicity / T cell extravasation / positive regulation of cellular extravasation / regulation of ruffle assembly / T cell antigen processing and presentation / T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell / membrane to membrane docking / adhesion of symbiont to host / establishment of endothelial barrier / cell adhesion mediated by integrin ...regulation of leukocyte mediated cytotoxicity / T cell extravasation / positive regulation of cellular extravasation / regulation of ruffle assembly / T cell antigen processing and presentation / T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell / membrane to membrane docking / adhesion of symbiont to host / establishment of endothelial barrier / cell adhesion mediated by integrin / heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules / leukocyte cell-cell adhesion / leukocyte migration / Interleukin-10 signaling /  immunological synapse / Integrin cell surface interactions / negative regulation of endothelial cell apoptotic process / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / cellular response to leukemia inhibitory factor / cellular response to glucose stimulus / cellular response to amyloid-beta / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / Interferon gamma signaling / transmembrane signaling receptor activity / immunological synapse / Integrin cell surface interactions / negative regulation of endothelial cell apoptotic process / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / cellular response to leukemia inhibitory factor / cellular response to glucose stimulus / cellular response to amyloid-beta / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / Interferon gamma signaling / transmembrane signaling receptor activity /  integrin binding / virus receptor activity / integrin binding / virus receptor activity /  signaling receptor activity / collagen-containing extracellular matrix / Interleukin-4 and Interleukin-13 signaling / receptor-mediated virion attachment to host cell / positive regulation of ERK1 and ERK2 cascade / signaling receptor activity / collagen-containing extracellular matrix / Interleukin-4 and Interleukin-13 signaling / receptor-mediated virion attachment to host cell / positive regulation of ERK1 and ERK2 cascade /  cell adhesion / cell adhesion /  membrane raft / external side of plasma membrane / membrane raft / external side of plasma membrane /  focal adhesion / focal adhesion /  cell surface / cell surface /  extracellular space / extracellular exosome / extracellular space / extracellular exosome /  membrane / membrane /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MIR / Resolution: 3.06 Å MIR / Resolution: 3.06 Å | |||||||||
 Authors Authors | Yang, Y. / Jun, C.D. / Liu, J.H. / Zhang, R. / Jochimiak, A. / Springer, T.A. / Wang, J.H. | |||||||||
 Citation Citation |  Journal: Mol.Cell / Year: 2004 Journal: Mol.Cell / Year: 2004Title: Structural basis for dimerization of ICAM-1 on the cell surface. Authors: Yang, Y. / Jun, C.D. / Liu, J.H. / Zhang, R. / Joachimiak, A. / Springer, T.A. / Wang, J.H. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1p53.cif.gz 1p53.cif.gz | 96.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1p53.ent.gz pdb1p53.ent.gz | 79.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1p53.json.gz 1p53.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/p5/1p53 https://data.pdbj.org/pub/pdb/validation_reports/p5/1p53 ftp://data.pdbj.org/pub/pdb/validation_reports/p5/1p53 ftp://data.pdbj.org/pub/pdb/validation_reports/p5/1p53 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 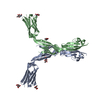
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  / ICAM-1 / Major group rhinovirus receptor / CD54 antigen / ICAM-1 / Major group rhinovirus receptor / CD54 antigenMass: 28829.492 Da / Num. of mol.: 2 / Fragment: ICAM-1 extracellular Domain 3-5, ecto-fragment Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: ICAM1 / Plasmid: pBJ5-GS / Cell line (production host): CHO.Lec3.2.8.1 / Production host: Homo sapiens (human) / Gene: ICAM1 / Plasmid: pBJ5-GS / Cell line (production host): CHO.Lec3.2.8.1 / Production host:   Cricetulus griseus (Chinese hamster) / References: UniProt: P05362 Cricetulus griseus (Chinese hamster) / References: UniProt: P05362#2: Sugar | ChemComp-NAG /  N-Acetylglucosamine N-Acetylglucosamine#3: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 5.5 Å3/Da / Density % sol: 77.5 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 5.6 Details: 15mg/ml protein, NH4H2PO4, 0.1 M Na-citrate, pH 5.6, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 113 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 1.008 Å / Beamline: 19-ID / Wavelength: 1.008 Å |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: May 11, 2000 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.008 Å / Relative weight: 1 : 1.008 Å / Relative weight: 1 |
| Reflection | Resolution: 3→20 Å / Num. all: 23462 / Num. obs: 23462 / % possible obs: 100 % / Observed criterion σ(F): -5 / Observed criterion σ(I): -5 / Redundancy: 7.7 % / Biso Wilson estimate: 65.2 Å2 / Rmerge(I) obs: 0.125 / Net I/σ(I): 10.8 |
| Reflection shell | Resolution: 3→3.11 Å / Redundancy: 7.8 % / Rmerge(I) obs: 0.415 / Mean I/σ(I) obs: 3.1 / Num. unique all: 2319 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MIR / Resolution: 3.06→20 Å / Isotropic thermal model: anisotropic / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber MIR / Resolution: 3.06→20 Å / Isotropic thermal model: anisotropic / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| |||||||||||||||||||||||||
| Displacement parameters | Biso mean: 35.184 Å2
| |||||||||||||||||||||||||
| Refine analyze |
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.06→20 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller









 PDBj
PDBj









