[English] 日本語
 Yorodumi
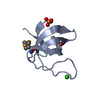
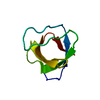
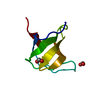
Yorodumi- PDB-1m3b: Solution structure of a circular form of the N-terminal SH3 domai... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1m3b | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution structure of a circular form of the N-terminal SH3 domain (A134C, E135G, R191G mutant) from oncogene protein c-Crk. | ||||||
 Components Components | Proto-oncogene C-crk | ||||||
 Keywords Keywords |  PROTEIN BINDING / SH3 / PROTEIN BINDING / SH3 /  SH3 DOMAIN / CIRCULAR PROTEIN / CYCLIZED PROTEIN / ADAPTOR PROTEIN SH3 DOMAIN / CIRCULAR PROTEIN / CYCLIZED PROTEIN / ADAPTOR PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationPTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / ARMS-mediated activation / MET activates RAP1 and RAC1 / MET receptor recycling / response to hepatocyte growth factor / cellular response to endothelin / helper T cell diapedesis / cerebellar neuron development / response to cholecystokinin / regulation of leukocyte migration ...PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / ARMS-mediated activation / MET activates RAP1 and RAC1 / MET receptor recycling / response to hepatocyte growth factor / cellular response to endothelin / helper T cell diapedesis / cerebellar neuron development / response to cholecystokinin / regulation of leukocyte migration / Downstream signal transduction / response to peptide / postsynaptic specialization assembly / protein phosphorylated amino acid binding / regulation of T cell migration / p130Cas linkage to MAPK signaling for integrins / reelin-mediated signaling pathway / regulation of dendrite development / positive regulation of skeletal muscle acetylcholine-gated channel clustering / response to yeast / Regulation of signaling by CBL / regulation of Rac protein signal transduction / Regulation of actin dynamics for phagocytic cup formation / negative regulation of wound healing / negative regulation of natural killer cell mediated cytotoxicity / negative regulation of cell motility / VEGFA-VEGFR2 Pathway / protein localization to membrane / cellular response to insulin-like growth factor stimulus / establishment of cell polarity /  regulation of GTPase activity / regulation of GTPase activity /  enzyme-linked receptor protein signaling pathway / positive regulation of smooth muscle cell migration / dendrite development / positive regulation of Rac protein signal transduction / regulation of cell adhesion mediated by integrin / cellular response to nitric oxide / ephrin receptor signaling pathway / signaling adaptor activity / positive regulation of substrate adhesion-dependent cell spreading / cellular response to transforming growth factor beta stimulus / enzyme-linked receptor protein signaling pathway / positive regulation of smooth muscle cell migration / dendrite development / positive regulation of Rac protein signal transduction / regulation of cell adhesion mediated by integrin / cellular response to nitric oxide / ephrin receptor signaling pathway / signaling adaptor activity / positive regulation of substrate adhesion-dependent cell spreading / cellular response to transforming growth factor beta stimulus /  insulin-like growth factor receptor binding / insulin-like growth factor receptor binding /  cytoskeletal protein binding / phosphotyrosine residue binding / cytoskeletal protein binding / phosphotyrosine residue binding /  SH2 domain binding / SH2 domain binding /  ephrin receptor binding / cell chemotaxis / ephrin receptor binding / cell chemotaxis /  protein tyrosine kinase binding / cellular response to nerve growth factor stimulus / hippocampus development / regulation of actin cytoskeleton organization / positive regulation of JNK cascade / protein tyrosine kinase binding / cellular response to nerve growth factor stimulus / hippocampus development / regulation of actin cytoskeleton organization / positive regulation of JNK cascade /  neuron migration / neuron migration /  neuromuscular junction / response to hydrogen peroxide / lipid metabolic process / neuromuscular junction / response to hydrogen peroxide / lipid metabolic process /  receptor tyrosine kinase binding / cerebral cortex development / receptor tyrosine kinase binding / cerebral cortex development /  SH3 domain binding / : / signaling receptor complex adaptor activity / SH3 domain binding / : / signaling receptor complex adaptor activity /  cell migration / cell migration /  actin cytoskeleton / protein-macromolecule adaptor activity / regulation of cell shape / actin cytoskeleton organization / actin cytoskeleton / protein-macromolecule adaptor activity / regulation of cell shape / actin cytoskeleton organization /  scaffold protein binding / cell population proliferation / protein domain specific binding / scaffold protein binding / cell population proliferation / protein domain specific binding /  ubiquitin protein ligase binding / ubiquitin protein ligase binding /  enzyme binding / enzyme binding /  signal transduction / protein-containing complex / signal transduction / protein-containing complex /  membrane / membrane /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  Simulated annealing, torsion angle dynamics Simulated annealing, torsion angle dynamics | ||||||
 Authors Authors | Schumann, F.H. / Varadan, R. / Tayakuniyil, P.P. / Hall, J.B. / Camarero, J.A. / Fushman, D. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain Authors: Schumann, F.H. / Varadan, R. / Tayakuniyil, P.P. / Hall, J.B. / Camarero, J.A. / Fushman, D. #1:  Journal: J.Mol.Biol. / Year: 2001 Journal: J.Mol.Biol. / Year: 2001Title: Rescuing a destabilized protein fold through backbone cyclization Authors: Camarero, J.A. / Fushman, D. / Sato, S. / Giriat, I. / Cowburn, D. / Raleigh, D.P. / Muir, T.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1m3b.cif.gz 1m3b.cif.gz | 361.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1m3b.ent.gz pdb1m3b.ent.gz | 311 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1m3b.json.gz 1m3b.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/m3/1m3b https://data.pdbj.org/pub/pdb/validation_reports/m3/1m3b ftp://data.pdbj.org/pub/pdb/validation_reports/m3/1m3b ftp://data.pdbj.org/pub/pdb/validation_reports/m3/1m3b | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 6831.592 Da / Num. of mol.: 1 / Fragment: N-TERMINAL SH3 DOMAIN (residues 134-191) / Mutation: R191G, E135G, A134C Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Gene: CRK / Plasmid: PGEX-6P-1 / Production host: Mus musculus (house mouse) / Gene: CRK / Plasmid: PGEX-6P-1 / Production host:   Escherichia coli (E. coli) / References: UniProt: Q64010 Escherichia coli (E. coli) / References: UniProt: Q64010 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques combined with 2D 1H-15N HSQC data. |
- Sample preparation
Sample preparation
| Details |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | Ionic strength: 100 mM NaCl / pH: 7.2 / Pressure: ambient / Temperature: 307 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | ||||||||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  Simulated annealing, torsion angle dynamics / Software ordinal: 1 Simulated annealing, torsion angle dynamics / Software ordinal: 1 Details: The structures are based on 1151 restraints, 1054 are NOE-derived distance contraints, 25 dihedral angle constraints, and 72 distance restraints from hydrogen bonds. Structures were ...Details: The structures are based on 1151 restraints, 1054 are NOE-derived distance contraints, 25 dihedral angle constraints, and 72 distance restraints from hydrogen bonds. Structures were calculated using program DYANA. No further refinement was performed. | ||||||||||||||||||||
| NMR representative | Selection criteria: lowest target function | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest target function Conformers calculated total number: 200 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller












 PDBj
PDBj





