[English] 日本語
 Yorodumi
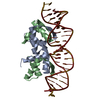
Yorodumi- PDB-1lmb: REFINED 1.8 ANGSTROM CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR-OP... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1lmb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | REFINED 1.8 ANGSTROM CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR-OPERATOR COMPLEX | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | TRANSCRIPTION/DNA /  PROTEIN-DNA COMPLEX / PROTEIN-DNA COMPLEX /  DOUBLE HELIX / TRANSCRIPTION-DNA COMPLEX DOUBLE HELIX / TRANSCRIPTION-DNA COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of viral latency / latency-replication decision / positive regulation of viral transcription / negative regulation of transcription by competitive promoter binding / core promoter sequence-specific DNA binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |   Enterobacteria phage lambda (virus) Enterobacteria phage lambda (virus) | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.8 Å X-RAY DIFFRACTION / Resolution: 1.8 Å | |||||||||
 Authors Authors | Beamer, L.J. / Pabo, C.O. | |||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1992 Journal: J.Mol.Biol. / Year: 1992Title: Refined 1.8 A crystal structure of the lambda repressor-operator complex. Authors: Beamer, L.J. / Pabo, C.O. #1:  Journal: Science / Year: 1991 Journal: Science / Year: 1991Title: The DNA Binding Arm of a Lambda Repressor: Critical Contacts from a Flexible Region Authors: Clarke, N.D. / Beamer, L.J. / Goldberg, H.R. / Berkower, C. / Pabo, C.O. #2:  Journal: Science / Year: 1990 Journal: Science / Year: 1990Title: Conserved Residues Make Similar Contacts in Two Repressor-Operator Complexes Authors: Pabo, C.O. / Aggarwal, A.K. / Jordan, S.R. / Beamer, L.J. / Obeysekare, U.R. / Harrison, S.C. #3:  Journal: J.Mol.Biol. / Year: 1983 Journal: J.Mol.Biol. / Year: 1983Title: Comparison of the Structures of Cro and Lambda Repressor Proteins from Bacteriophage Lambda Authors: Ohlendorf, D.H. / Anderson, W.F. / Lewis, M. / Pabo, C.O. / Matthews, B.W. #4:  Journal: Cold Spring Harbor Symp.Quant.Biol. / Year: 1983 Journal: Cold Spring Harbor Symp.Quant.Biol. / Year: 1983Title: Structure of the Operator-Binding Domain of Bacteriophage Lambda Repressor. Implications for DNA Recognition and Gene Regulation Authors: Lewis, M. / Jeffrey, A. / Wang, J. / Ladner, R. / Ptashne, M. / Pabo, C.O. #5:  Journal: Nature / Year: 1982 Journal: Nature / Year: 1982Title: Homology Among DNA-Binding Proteins Suggests Use of a Conserved Super-Secondary Structure Authors: Sauer, R.T. / Yocum, R.R. / Doolittle, R.F. / Lewis, M. / Pabo, C.O. #6:  Journal: Nature / Year: 1982 Journal: Nature / Year: 1982Title: The Operator-Binding Domain of Lambda Repressor. Structure and DNA Recognition Authors: Pabo, C.O. / Lewis, M. #7:  Journal: Nature / Year: 1982 Journal: Nature / Year: 1982Title: The N-Terminal Arms of Lambda Repressor Wrap Around the Operator DNA Authors: Pabo, C.O. / Krovatin, W. / Jeffrey, A. / Sauer, R.T. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1lmb.cif.gz 1lmb.cif.gz | 72.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1lmb.ent.gz pdb1lmb.ent.gz | 51 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1lmb.json.gz 1lmb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lm/1lmb https://data.pdbj.org/pub/pdb/validation_reports/lm/1lmb ftp://data.pdbj.org/pub/pdb/validation_reports/lm/1lmb ftp://data.pdbj.org/pub/pdb/validation_reports/lm/1lmb | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 6158.004 Da / Num. of mol.: 1 / Source method: obtained synthetically | ||
|---|---|---|---|
| #2: DNA chain | Mass: 6108.967 Da / Num. of mol.: 1 / Source method: obtained synthetically | ||
| #3: Protein | Mass: 10136.613 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   Enterobacteria phage lambda (virus) / Genus: Lambda-like viruses / References: UniProt: P03034 Enterobacteria phage lambda (virus) / Genus: Lambda-like viruses / References: UniProt: P03034#4: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.21 Å3/Da / Density % sol: 44.29 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | Method: vapor diffusion / pH: 7 / Details: pH 7.00, VAPOR DIFFUSION | ||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 20 ℃ / Method: vapor diffusion, hanging dropDetails: taken from Jordan, S.R. et al.(1985). Science, 230, 1383-1385. | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 258 K |
|---|---|
| Detector | Type: UCSD MARK II / Detector: AREA DETECTOR |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Highest resolution: 1.8 Å / Num. obs: 28193 / Observed criterion σ(F): 2 |
| Reflection | *PLUS Highest resolution: 1.8 Å / Lowest resolution: 8 Å / Num. measured all: 101275 / Rmerge(I) obs: 0.052 |
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.8→8 Å / σ(F): 2 /
| ||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→8 Å
| ||||||||||||
| Refinement | *PLUS Highest resolution: 1.8 Å / Lowest resolution: 8 Å / Num. reflection obs: 25252 / Rfactor obs: 0.189 | ||||||||||||
| Solvent computation | *PLUS | ||||||||||||
| Displacement parameters | *PLUS Biso mean: 25.2 Å2 |
 Movie
Movie Controller
Controller









 PDBj
PDBj








































