[English] 日本語
 Yorodumi
Yorodumi- PDB-1lfu: NMR Solution Structure of the Extended PBX Homeodomain Bound to DNA -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1lfu | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR Solution Structure of the Extended PBX Homeodomain Bound to DNA | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  TRANSCRIPTION / TRANSCRIPTION /  protein-dna complex protein-dna complex | ||||||
| Function / homology |  Function and homology information Function and homology information urogenital system development / natural killer cell differentiation / proximal/distal pattern formation / embryonic skeletal system development / urogenital system development / natural killer cell differentiation / proximal/distal pattern formation / embryonic skeletal system development /  sex differentiation / sex differentiation /  eye development / steroid biosynthetic process / embryonic limb morphogenesis / embryonic hemopoiesis / anterior/posterior pattern specification ... eye development / steroid biosynthetic process / embryonic limb morphogenesis / embryonic hemopoiesis / anterior/posterior pattern specification ... urogenital system development / natural killer cell differentiation / proximal/distal pattern formation / embryonic skeletal system development / urogenital system development / natural killer cell differentiation / proximal/distal pattern formation / embryonic skeletal system development /  sex differentiation / sex differentiation /  eye development / steroid biosynthetic process / embryonic limb morphogenesis / embryonic hemopoiesis / anterior/posterior pattern specification / branching involved in ureteric bud morphogenesis / adrenal gland development / positive regulation of stem cell proliferation / negative regulation of neuron differentiation / eye development / steroid biosynthetic process / embryonic limb morphogenesis / embryonic hemopoiesis / anterior/posterior pattern specification / branching involved in ureteric bud morphogenesis / adrenal gland development / positive regulation of stem cell proliferation / negative regulation of neuron differentiation /  regulation of ossification / neuron development / embryonic organ development / spleen development / positive regulation of G2/M transition of mitotic cell cycle / transcription corepressor binding / thymus development / stem cell proliferation / regulation of ossification / neuron development / embryonic organ development / spleen development / positive regulation of G2/M transition of mitotic cell cycle / transcription corepressor binding / thymus development / stem cell proliferation /  transcription coregulator binding / RNA polymerase II transcription regulatory region sequence-specific DNA binding / animal organ morphogenesis / transcription coregulator binding / RNA polymerase II transcription regulatory region sequence-specific DNA binding / animal organ morphogenesis /  brain development / RNA polymerase II transcription regulator complex / G2/M transition of mitotic cell cycle / sequence-specific double-stranded DNA binding / regulation of cell population proliferation / DNA-binding transcription activator activity, RNA polymerase II-specific / DNA-binding transcription factor binding / cell population proliferation / brain development / RNA polymerase II transcription regulator complex / G2/M transition of mitotic cell cycle / sequence-specific double-stranded DNA binding / regulation of cell population proliferation / DNA-binding transcription activator activity, RNA polymerase II-specific / DNA-binding transcription factor binding / cell population proliferation /  transcription regulator complex / sequence-specific DNA binding / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / transcription regulator complex / sequence-specific DNA binding / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II /  DNA binding / DNA binding /  nucleoplasm / nucleoplasm /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | ||||||
| Method |  SOLUTION NMR / dynamical annealing SOLUTION NMR / dynamical annealing | ||||||
 Authors Authors | Sprules, T. / Green, N. / Featherstone, M. / Gehring, K. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2003 Journal: J.Biol.Chem. / Year: 2003Title: Lock and Key Binding of the HOX YPWM Peptide to the PBX Homeodomain Authors: Sprules, T. / Green, N. / Featherstone, M. / Gehring, K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1lfu.cif.gz 1lfu.cif.gz | 869.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1lfu.ent.gz pdb1lfu.ent.gz | 753.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1lfu.json.gz 1lfu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lf/1lfu https://data.pdbj.org/pub/pdb/validation_reports/lf/1lfu ftp://data.pdbj.org/pub/pdb/validation_reports/lf/1lfu ftp://data.pdbj.org/pub/pdb/validation_reports/lf/1lfu | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 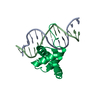
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 4256.766 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: DNA chain | Mass: 4305.804 Da / Num. of mol.: 1 / Source method: obtained synthetically |
| #3: Protein |  / PBX / PRE-B-CELL LEUKEMIA TRANSCRIPTION FACTOR-1 / PBX / PRE-B-CELL LEUKEMIA TRANSCRIPTION FACTOR-1Mass: 9577.833 Da / Num. of mol.: 1 / Fragment: homeodomain AND conserved C-terminus / Mutation: C42S Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Gene: PBX1 / Plasmid: pET 11a / Production host: Mus musculus (house mouse) / Gene: PBX1 / Plasmid: pET 11a / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21(DE3)Gold / References: UniProt: P41778 Escherichia coli (E. coli) / Strain (production host): BL21(DE3)Gold / References: UniProt: P41778 |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
| ||||||||||||||||||||||||||||
Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: dynamical annealing / Software ordinal: 1 Details: Structures based on a total of 2747 restraints: 2508 NOE-derived distance restraints, 84 hydrogen bonds, 68 dihedral angle and J-coupling restraints and 87 residual dipolar couplings. | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: lowest energy with acceptable geometry Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller












 PDBj
PDBj






































