[English] 日本語
 Yorodumi
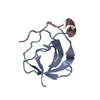
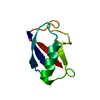
Yorodumi- PDB-1ka5: Refined Solution Structure of Histidine Containing Phosphocarrier... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ka5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Refined Solution Structure of Histidine Containing Phosphocarrier Protein from Staphyloccocus aureus | |||||||||
 Components Components | PHOSPHOCARRIER PROTEIN HPR | |||||||||
 Keywords Keywords | LIGAND TRANSPORT / Open Faced beta-Sandwich /  Structural Proteomics in Europe / SPINE / Structural Proteomics in Europe / SPINE /  Structural Genomics Structural Genomics | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphoenolpyruvate-dependent sugar phosphotransferase system /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Staphylococcus aureus (bacteria) Staphylococcus aureus (bacteria) | |||||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing simulated annealing | |||||||||
 Authors Authors | Maurer, T. / Meier, S. / Hengstenberg, W. / Kalbitzer, H.R. / Structural Proteomics in Europe (SPINE) | |||||||||
 Citation Citation |  Journal: J.Bacteriol. / Year: 2004 Journal: J.Bacteriol. / Year: 2004Title: High-resolution structure of the histidine-containing phosphocarrier protein (HPr) from Staphylococcus aureus and characterization of its interaction with the bifunctional HPr kinase/phosphorylase Authors: Maurer, T. / Meier, S. / Kachel, N. / Munte, C.E. / Hasenbein, S. / Koch, B. / Hengstenberg, W. / Kalbitzer, H.R. #1:  Journal: Eur.J.Biochem. / Year: 1993 Journal: Eur.J.Biochem. / Year: 1993Title: The Solution Structure of the Histidine Containing Phosphocarrier Protein (HPr) from Staphylococcus aureus as Determined by Two-Dimensional 1H-NMR Spectroscopy Authors: Kalbitzer, H.R. / Hengstenberg, W. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ka5.cif.gz 1ka5.cif.gz | 413.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ka5.ent.gz pdb1ka5.ent.gz | 358.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ka5.json.gz 1ka5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ka/1ka5 https://data.pdbj.org/pub/pdb/validation_reports/ka/1ka5 ftp://data.pdbj.org/pub/pdb/validation_reports/ka/1ka5 ftp://data.pdbj.org/pub/pdb/validation_reports/ka/1ka5 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 9504.689 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Staphylococcus aureus (bacteria) / References: UniProt: P0A0E3 Staphylococcus aureus (bacteria) / References: UniProt: P0A0E3 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 2mM HPr / Solvent system: 95% H2O, 5% 2H2O |
|---|---|
| Sample conditions | Ionic strength: 0 / pH: 7.4 / Pressure: ambinet / Temperature: 298 K |
Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | |||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing / Software ordinal: 1 simulated annealing / Software ordinal: 1 Details: 1883 distance and angle restraints and hydrogen bonding patterns | ||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 440 / Conformers submitted total number: 16 |
 Movie
Movie Controller
Controller










 PDBj
PDBj