+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1gqf | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of human procaspase-7 | ||||||
 Components Components | Caspase-7 Caspase 7 Caspase 7 | ||||||
 Keywords Keywords |  HYDROLASE / HYDROLASE /  CASPASE-7 / CASPASE-7 /  APOPTOSIS / APOPTOSIS /  ZYMOGEN ZYMOGEN | ||||||
| Function / homology |  Function and homology information Function and homology information caspase-7 / lymphocyte apoptotic process / positive regulation of plasma membrane repair / cellular response to staurosporine / cysteine-type endopeptidase activity involved in execution phase of apoptosis / SMAC, XIAP-regulated apoptotic response / Activation of caspases through apoptosome-mediated cleavage / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes / cysteine-type endopeptidase activity involved in apoptotic process ... caspase-7 / lymphocyte apoptotic process / positive regulation of plasma membrane repair / cellular response to staurosporine / cysteine-type endopeptidase activity involved in execution phase of apoptosis / SMAC, XIAP-regulated apoptotic response / Activation of caspases through apoptosome-mediated cleavage / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes / cysteine-type endopeptidase activity involved in apoptotic process ... caspase-7 / lymphocyte apoptotic process / positive regulation of plasma membrane repair / cellular response to staurosporine / cysteine-type endopeptidase activity involved in execution phase of apoptosis / SMAC, XIAP-regulated apoptotic response / Activation of caspases through apoptosome-mediated cleavage / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes / cysteine-type endopeptidase activity involved in apoptotic process / fibroblast apoptotic process / execution phase of apoptosis / Apoptotic cleavage of cellular proteins / protein maturation / Caspase-mediated cleavage of cytoskeletal proteins / cysteine-type peptidase activity / response to UV / striated muscle cell differentiation / protein catabolic process / protein processing / caspase-7 / lymphocyte apoptotic process / positive regulation of plasma membrane repair / cellular response to staurosporine / cysteine-type endopeptidase activity involved in execution phase of apoptosis / SMAC, XIAP-regulated apoptotic response / Activation of caspases through apoptosome-mediated cleavage / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes / cysteine-type endopeptidase activity involved in apoptotic process / fibroblast apoptotic process / execution phase of apoptosis / Apoptotic cleavage of cellular proteins / protein maturation / Caspase-mediated cleavage of cytoskeletal proteins / cysteine-type peptidase activity / response to UV / striated muscle cell differentiation / protein catabolic process / protein processing /  heart development / heart development /  peptidase activity / neuron apoptotic process / cellular response to lipopolysaccharide / aspartic-type endopeptidase activity / defense response to bacterium / cysteine-type endopeptidase activity / apoptotic process / peptidase activity / neuron apoptotic process / cellular response to lipopolysaccharide / aspartic-type endopeptidase activity / defense response to bacterium / cysteine-type endopeptidase activity / apoptotic process /  proteolysis / proteolysis /  extracellular space / extracellular space /  RNA binding / RNA binding /  nucleoplasm / nucleoplasm /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.9 Å MOLECULAR REPLACEMENT / Resolution: 2.9 Å | ||||||
 Authors Authors | Riedl, S. / Bode, W. / Fuentes-Prior, P. | ||||||
 Citation Citation |  Journal: Proc. Natl. Acad. Sci. U.S.A. / Year: 2001 Journal: Proc. Natl. Acad. Sci. U.S.A. / Year: 2001Title: Structural basis for the activation of human procaspase-7. Authors: Riedl, S.J. / Fuentes-Prior, P. / Renatus, M. / Kairies, N. / Krapp, S. / Huber, R. / Salvesen, G.S. / Bode, W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1gqf.cif.gz 1gqf.cif.gz | 121.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1gqf.ent.gz pdb1gqf.ent.gz | 93.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1gqf.json.gz 1gqf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gq/1gqf https://data.pdbj.org/pub/pdb/validation_reports/gq/1gqf ftp://data.pdbj.org/pub/pdb/validation_reports/gq/1gqf ftp://data.pdbj.org/pub/pdb/validation_reports/gq/1gqf | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 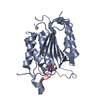 1f1jS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  Caspase 7 / CASP-7 / Apoptotic protease Mch-3 / CMH-1 / ICE-like apoptotic protease 3 / ICE-LAP3 Caspase 7 / CASP-7 / Apoptotic protease Mch-3 / CMH-1 / ICE-like apoptotic protease 3 / ICE-LAP3Mass: 30283.334 Da / Num. of mol.: 2 / Mutation: C187A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: CASP7, MCH3 / Production host: Homo sapiens (human) / Gene: CASP7, MCH3 / Production host:   Escherichia coli (E. coli) / References: UniProt: P55210, Escherichia coli (E. coli) / References: UniProt: P55210,  caspase-7 caspase-7#2: Chemical | ChemComp-SO4 /  Sulfate Sulfate#3: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.55 Å3/Da / Density % sol: 60 % | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion / pH: 5.6 Details: 50 MM MES, PH 5.6, 10 MM MAGNESIUM CHLORIDE, 100 MM AMMONIUM SULFATE, 20% (W/V) PEG 8000 | |||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 20 ℃ / Method: vapor diffusion, sitting drop / pH: 5.6 | |||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: MPG/DESY, HAMBURG SYNCHROTRON / Site: MPG/DESY, HAMBURG  / Beamline: BW6 / Wavelength: 1.05 / Beamline: BW6 / Wavelength: 1.05 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Jul 23, 2001 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.05 Å / Relative weight: 1 : 1.05 Å / Relative weight: 1 |
| Reflection | Resolution: 2.9→59.76 Å / Num. obs: 19487 / % possible obs: 99.1 % / Observed criterion σ(I): 2 / Redundancy: 3.4 % / Biso Wilson estimate: 74.2 Å2 / Rmerge(I) obs: 0.068 |
| Reflection shell | Resolution: 2.9→3.06 Å / Redundancy: 3.1 % / Rmerge(I) obs: 0.386 / % possible all: 99.1 |
| Reflection | *PLUS Num. measured all: 180534 |
| Reflection shell | *PLUS % possible obs: 99.1 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1F1J Resolution: 2.9→21.73 Å / Rfactor Rfree error: 0.009 / Data cutoff high absF: 1166291.1 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 Details: COMPETITION FOR THE LIMITED INTERMONOMER SPACE (THE "CENTRAL CAVITY") BY THE TWO INTERDOMAIN LINKERS MANIFESTS ITSELF IN POOR OR AMBIGOUS ELECTRON DENSITY FOR RESIDUES N-TERMINAL OF ILE321. ...Details: COMPETITION FOR THE LIMITED INTERMONOMER SPACE (THE "CENTRAL CAVITY") BY THE TWO INTERDOMAIN LINKERS MANIFESTS ITSELF IN POOR OR AMBIGOUS ELECTRON DENSITY FOR RESIDUES N-TERMINAL OF ILE321. ALTERNATE CONFORMATIONS, HOWEVER, ARE NOT SEPARABLE AT THE CURRENT RESOLUTION. ELECTRON DENSITY IS ALSO POOR OR MISSING FOR RESIDUES THR288 - LYS320 (THE INTERDOMAIN LINKER), GLY336 - GLY346 (THE SUBSTRATE BINDING LOOP), AS WELL AS FOR THE INSERTION LOOP GLU381-GLU382. RESIDUES N-TERMINAL OF PRO149 AND THE C-TERMINAL HISTIDINE TAG ARE FLEXIBLY DISORDERED.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 210.532 Å2 / ksol: 0.237854 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 80.7 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.9→21.73 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.9→3.08 Å / Rfactor Rfree error: 0.033 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Version: 1.1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 59.76 Å / % reflection Rfree: 5 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor obs: 0.313 |
 Movie
Movie Controller
Controller














 PDBj
PDBj






