[English] 日本語
 Yorodumi
Yorodumi- PDB-1goy: HYDROLASE(ENDORIBONUCLEASE)RIBONUCLEASE BI(G SPECIFIC ENDONUCLEAS... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1goy | ||||||
|---|---|---|---|---|---|---|---|
| Title | HYDROLASE(ENDORIBONUCLEASE)RIBONUCLEASE BI(G SPECIFIC ENDONUCLEASE) (E.C.3.1.27.-) COMPLEXED WITH GUANOSINE-3'-PHOSPHATE (3'-GMP) | ||||||
 Components Components | RIBONUCLEASE | ||||||
 Keywords Keywords |  HYDROLASE / HYDROLASE /  ENDORIBONUCLEASE / ENDORIBONUCLEASE /  NUCLEASE NUCLEASE | ||||||
| Function / homology |  Function and homology information Function and homology information Hydrolases; Acting on ester bonds; Endoribonucleases producing 3'-phosphomonoesters / RNA endonuclease activity / Hydrolases; Acting on ester bonds; Endoribonucleases producing 3'-phosphomonoesters / RNA endonuclease activity /  RNA binding / extracellular region RNA binding / extracellular regionSimilarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MIR / Resolution: 2 Å MIR / Resolution: 2 Å | ||||||
 Authors Authors | Polyakov, K.M. / Lebedev, A.A. / Pavlovsky, A.G. / Sanishvili, R.G. / Dodson, G.G. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2002 Journal: Acta Crystallogr.,Sect.D / Year: 2002Title: The Structure of Substrate-Free Microbial Ribonuclease Binase and of its Complexes with 3'Gmp and Sulfate Ions Authors: Polyakov, K.M. / Lebedev, A.A. / Okorokov, A.L. / Panov, K.I. / Schulga, A.A. / Pavlovsky, A.G. / Karpeisky, M.Y.A. / Dodson, G.G. #1: Journal: Trends Biochem.Sci. / Year: 1990 Title: Comparison of Active Sites of Some Microbial Ribonucleases: Structural Basis for Guanylic Specificity Authors: Sevcik, J. / Sanishvili, R.G. / Pavlovsky, A.G. / Polyakov, K.M. #2:  Journal: Kristallografiya(Russian) / Year: 1989 Journal: Kristallografiya(Russian) / Year: 1989Title: The Crystall Structure of Bacterial Ribonuclease Binase Authors: Pavlovsky, A.G. / Sanishvili, R.G. / Borisova, S.N. / Strokopytov, B.V. / Vagin, A.A. / Chepurnova, N.K. / Vainshtein, B.K. #3: Journal: FEBS Lett. / Year: 1983 Title: Three-Dimensional Structure of Ribonuclese from Balillus Intermedius 7P at 3A Resolution Authors: Pavlovsky, A.G. / Vagin, A.A. / Vainshtein, B.K. / Chepurnova, N.K. / Karpeisky, M.Y. #4:  Journal: Trends Biochem.Sci. / Year: 1983 Journal: Trends Biochem.Sci. / Year: 1983Title: The Structural and Sequence Homology of Family of Microbial Ribonucleases Authors: Hill, C. / Dodson, G. / Heinemann, U. / Saenger, W. / Mitsui, Y. / Nakamura, K. / Borisov, S. / Tischenko, G. / Polyakov, K. / Pavlovsky, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1goy.cif.gz 1goy.cif.gz | 61.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1goy.ent.gz pdb1goy.ent.gz | 45.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1goy.json.gz 1goy.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/go/1goy https://data.pdbj.org/pub/pdb/validation_reports/go/1goy ftp://data.pdbj.org/pub/pdb/validation_reports/go/1goy ftp://data.pdbj.org/pub/pdb/validation_reports/go/1goy | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 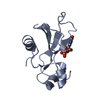
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  / BINASE / G SPECIFIC ENDONUCLEASE / BINASE / G SPECIFIC ENDONUCLEASEMass: 12974.492 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: MICROORGANISM / Source: (natural)  #2: Chemical | ChemComp-3GP / |  Guanosine monophosphate Guanosine monophosphate#3: Chemical |  Sulfate Sulfate#4: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.5 Å3/Da / Density % sol: 50.75 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | pH: 7 / Details: pH 7.00 | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, sitting drop | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 287 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Wavelength: 1.5418 ROTATING ANODE / Wavelength: 1.5418 |
| Detector | Type: SYTNEX P21 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2→10 Å / Num. obs: 14982 / % possible obs: 99 % / Redundancy: 2 % |
| Reflection | *PLUS Highest resolution: 2 Å / Lowest resolution: 24.9 Å / Num. obs: 15152 / % possible obs: 82.9 % / Num. measured all: 15152 |
| Reflection shell | *PLUS Highest resolution: 2 Å / Lowest resolution: 2.05 Å / % possible obs: 34.2 % / Mean I/σ(I) obs: 1.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MIR / Resolution: 2→10 Å / Cor.coef. Fo:Fc: 0.951 / SU B: 3.682 / SU ML: 0.095 / Cross valid method: THROUGHOUT / ESU R: 0.192 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: NONE MIR / Resolution: 2→10 Å / Cor.coef. Fo:Fc: 0.951 / SU B: 3.682 / SU ML: 0.095 / Cross valid method: THROUGHOUT / ESU R: 0.192 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: NONE
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL PLUS MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller
















 PDBj
PDBj