+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4231 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | C1-IgG1 complex on liposomes | |||||||||||||||||||||
 Map data Map data | C1-IgG1 complex observed on liposomes | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcomplement component C1 complex / complement component C1q complex / negative regulation of macrophage differentiation / synapse pruning / negative regulation of granulocyte differentiation / vertebrate eye-specific patterning / complement-mediated synapse pruning / collagen trimer /  complement activation / Classical antibody-mediated complement activation ...complement component C1 complex / complement component C1q complex / negative regulation of macrophage differentiation / synapse pruning / negative regulation of granulocyte differentiation / vertebrate eye-specific patterning / complement-mediated synapse pruning / collagen trimer / complement activation / Classical antibody-mediated complement activation ...complement component C1 complex / complement component C1q complex / negative regulation of macrophage differentiation / synapse pruning / negative regulation of granulocyte differentiation / vertebrate eye-specific patterning / complement-mediated synapse pruning / collagen trimer /  complement activation / Classical antibody-mediated complement activation / neuron remodeling / Initial triggering of complement / complement activation / Classical antibody-mediated complement activation / neuron remodeling / Initial triggering of complement /  immunoglobulin complex / immunoglobulin complex /  complement activation, classical pathway / complement activation, classical pathway /  Regulation of Complement cascade / astrocyte activation / synapse organization / microglial cell activation / cell-cell signaling / Regulation of Complement cascade / astrocyte activation / synapse organization / microglial cell activation / cell-cell signaling /  amyloid-beta binding / postsynapse / collagen-containing extracellular matrix / blood microparticle / amyloid-beta binding / postsynapse / collagen-containing extracellular matrix / blood microparticle /  adaptive immune response / adaptive immune response /  immune response / immune response /  innate immune response / innate immune response /  synapse / synapse /  extracellular space / extracellular region / extracellular space / extracellular region /  plasma membrane plasma membraneSimilarity search - Function | |||||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
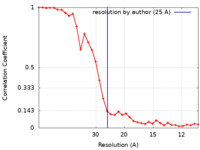
| Method | subtomogram averaging /  cryo EM / Resolution: 25.0 Å cryo EM / Resolution: 25.0 Å | |||||||||||||||||||||
 Authors Authors | Howes SC / Koning RI / de Jong RN / Beurskens FJ / Koster AJ / Sharp TH | |||||||||||||||||||||
| Funding support |  Netherlands, 6 items Netherlands, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement. Authors: Deniz Ugurlar / Stuart C Howes / Bart-Jan de Kreuk / Roman I Koning / Rob N de Jong / Frank J Beurskens / Janine Schuurman / Abraham J Koster / Thomas H Sharp / Paul W H I Parren / Piet Gros /  Abstract: Danger patterns on microbes or damaged host cells bind and activate C1, inducing innate immune responses and clearance through the complement cascade. How these patterns trigger complement initiation ...Danger patterns on microbes or damaged host cells bind and activate C1, inducing innate immune responses and clearance through the complement cascade. How these patterns trigger complement initiation remains elusive. Here, we present cryo-electron microscopy analyses of C1 bound to monoclonal antibodies in which we observed heterogeneous structures of single and clustered C1-immunoglobulin G1 (IgG1) hexamer complexes. Distinct C1q binding sites are observed on the two Fc-CH2 domains of each IgG molecule. These are consistent with known interactions and also reveal additional interactions, which are supported by functional IgG1-mutant analysis. Upon antibody binding, the C1q arms condense, inducing rearrangements of the C1rs proteases and tilting C1q's cone-shaped stalk. The data suggest that C1r may activate C1s within single, strained C1 complexes or between neighboring C1 complexes on surfaces. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4231.map.gz emd_4231.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4231-v30.xml emd-4231-v30.xml emd-4231.xml emd-4231.xml | 27.3 KB 27.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4231_fsc.xml emd_4231_fsc.xml | 3.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_4231.png emd_4231.png | 82.6 KB | ||
| Others |  emd_4231_additional_1.map.gz emd_4231_additional_1.map.gz emd_4231_additional_2.map.gz emd_4231_additional_2.map.gz emd_4231_additional_3.map.gz emd_4231_additional_3.map.gz emd_4231_additional_4.map.gz emd_4231_additional_4.map.gz emd_4231_half_map_1.map.gz emd_4231_half_map_1.map.gz emd_4231_half_map_2.map.gz emd_4231_half_map_2.map.gz | 10.8 MB 1.7 MB 10.7 MB 10.7 MB 2.2 MB 2.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4231 http://ftp.pdbj.org/pub/emdb/structures/EMD-4231 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4231 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4231 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4231.map.gz / Format: CCP4 / Size: 2.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4231.map.gz / Format: CCP4 / Size: 2.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C1-IgG1 complex observed on liposomes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Class 1 of neighboring C1-IgG1 complexes
| File | emd_4231_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class 1 of neighboring C1-IgG1 complexes | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: C1-IgG1 complex observed on liposomes refined using mask...
| File | emd_4231_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C1-IgG1 complex observed on liposomes refined using mask that excluded the membrane and Fab regions | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Class 2 of neighboring C1-IgG1 complexes
| File | emd_4231_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class 2 of neighboring C1-IgG1 complexes | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Class 3 of neighboring C1-IgG1 complexes
| File | emd_4231_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class 3 of neighboring C1-IgG1 complexes | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of C1-IgG1 complex observed on liposomes
| File | emd_4231_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of C1-IgG1 complex observed on liposomes | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of C1-IgG1 complex observed on liposomes
| File | emd_4231_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of C1-IgG1 complex observed on liposomes | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : C1-IgG1 complex on liposomes
| Entire | Name: C1-IgG1 complex on liposomes |
|---|---|
| Components |
|
-Supramolecule #1: C1-IgG1 complex on liposomes
| Supramolecule | Name: C1-IgG1 complex on liposomes / type: complex / ID: 1 / Parent: 0 Details: Liposomes containing di-nitrophenyl haptens were incubated with monoclonal antibodies and C1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 700 KDa |
-Supramolecule #2: IgG1 antibodies
| Supramolecule | Name: IgG1 antibodies / type: complex / ID: 2 / Parent: 1 Details: Monoclonal IgG1 recombinantly expressed in HEK293 FreeStyle cells, purified and bound to liposomes. |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK293 Homo sapiens (human) / Recombinant cell: HEK293 |
-Supramolecule #3: C1 complex
| Supramolecule | Name: C1 complex / type: complex / ID: 3 / Parent: 1 Details: C1 complex purified from human serum containing C1q, C1r, and C1s. Map from focused alignment and classification. |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 96 % / Chamber temperature: 294 K / Instrument: LEICA EM GP Details: 3 microlitres applied and incubated for 30 seconds, blot for 1 second before plunging. | ||||||||||||
| Details | Extruded liposomes were incubated with IgG1 and C1. Gold fiducials were added just before applying to grids. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus min: 0.3 µm / Nominal magnification: 53000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus min: 0.3 µm / Nominal magnification: 53000 |
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: GIF Quantum LS |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-6 / Average electron dose: 1.2 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z
Z Y
Y X
X