+ Open data
Open data
- Basic information
Basic information
| Entry | 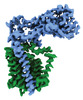 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Periplasmic map | |||||||||
 Map data Map data | Periplasmic map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Peptidoglycan / Peptidoglycan /  glycosyltransferase / glycosyltransferase /  enzyme / enzyme /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.95 Å cryo EM / Resolution: 2.95 Å | |||||||||
 Authors Authors | Nygaard R / Mancia F | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex Authors: Nygaard R / Graham CLB / Belcher Dufrisne M / Colburn JD / Pepe J / Hydorn MA / Corradi S / Brown CM / Ashraf KU / Vickery ON / Briggs NS / Deering JJ / Kloss B / Botta B / Clarke OB / ...Authors: Nygaard R / Graham CLB / Belcher Dufrisne M / Colburn JD / Pepe J / Hydorn MA / Corradi S / Brown CM / Ashraf KU / Vickery ON / Briggs NS / Deering JJ / Kloss B / Botta B / Clarke OB / Columbus L / Dworkin J / Stansfeld PJ / Roper DI / Mancia F | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41304.map.gz emd_41304.map.gz | 122.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41304-v30.xml emd-41304-v30.xml emd-41304.xml emd-41304.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41304.png emd_41304.png | 118.4 KB | ||
| Others |  emd_41304_half_map_1.map.gz emd_41304_half_map_1.map.gz emd_41304_half_map_2.map.gz emd_41304_half_map_2.map.gz | 225.3 MB 225.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41304 http://ftp.pdbj.org/pub/emdb/structures/EMD-41304 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41304 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41304 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41304.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41304.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Periplasmic map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_41304_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_41304_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RodA-PBP2
| Entire | Name: RodA-PBP2 |
|---|---|
| Components |
|
-Supramolecule #1: RodA-PBP2
| Supramolecule | Name: RodA-PBP2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / Details: Nanodisc were formed using MSP1E3D1 and POPG lipid |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 111.803 KDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.66 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 11120 / Average exposure time: 2.5 sec. / Average electron dose: 58.5 e/Å2 |
- Image processing
Image processing
| Particle selection | Number selected: 4415933 |
|---|---|
| Startup model | Type of model: OTHER |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.95 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2.12 and 3.2) / Software - details: Local refinement / Number images used: 104553 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X


















