[English] 日本語
 Yorodumi
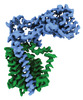
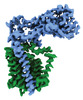
Yorodumi- EMDB-41299: Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 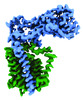 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Peptidoglycan / Peptidoglycan /  glycosyltransferase / glycosyltransferase /  enzyme / enzyme /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information peptidoglycan glycosyltransferase / peptidoglycan glycosyltransferase /  peptidoglycan glycosyltransferase activity / peptidoglycan glycosyltransferase activity /  serine-type D-Ala-D-Ala carboxypeptidase / serine-type D-Ala-D-Ala carboxypeptidase /  serine-type D-Ala-D-Ala carboxypeptidase activity / serine-type D-Ala-D-Ala carboxypeptidase activity /  acyltransferase activity / acyltransferase activity /  glycosyltransferase activity / glycosyltransferase activity /  penicillin binding / peptidoglycan biosynthetic process / cell wall organization / regulation of cell shape ... penicillin binding / peptidoglycan biosynthetic process / cell wall organization / regulation of cell shape ... peptidoglycan glycosyltransferase / peptidoglycan glycosyltransferase /  peptidoglycan glycosyltransferase activity / peptidoglycan glycosyltransferase activity /  serine-type D-Ala-D-Ala carboxypeptidase / serine-type D-Ala-D-Ala carboxypeptidase /  serine-type D-Ala-D-Ala carboxypeptidase activity / serine-type D-Ala-D-Ala carboxypeptidase activity /  acyltransferase activity / acyltransferase activity /  glycosyltransferase activity / glycosyltransferase activity /  penicillin binding / peptidoglycan biosynthetic process / cell wall organization / regulation of cell shape / penicillin binding / peptidoglycan biosynthetic process / cell wall organization / regulation of cell shape /  cell division / cell division /  proteolysis / proteolysis /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Nygaard R / Mancia F | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex Authors: Nygaard R / Graham CLB / Belcher Dufrisne M / Colburn JD / Pepe J / Hydorn MA / Corradi S / Brown CM / Ashraf KU / Vickery ON / Briggs NS / Deering JJ / Kloss B / Botta B / Clarke OB / ...Authors: Nygaard R / Graham CLB / Belcher Dufrisne M / Colburn JD / Pepe J / Hydorn MA / Corradi S / Brown CM / Ashraf KU / Vickery ON / Briggs NS / Deering JJ / Kloss B / Botta B / Clarke OB / Columbus L / Dworkin J / Stansfeld PJ / Roper DI / Mancia F | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41299.map.gz emd_41299.map.gz | 4.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41299-v30.xml emd-41299-v30.xml emd-41299.xml emd-41299.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41299.png emd_41299.png | 494.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41299 http://ftp.pdbj.org/pub/emdb/structures/EMD-41299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41299 | HTTPS FTP |
-Related structure data
| Related structure data |  8tj3MC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41299.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41299.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : RodA-PBP2
| Entire | Name: RodA-PBP2 |
|---|---|
| Components |
|
-Supramolecule #1: RodA-PBP2
| Supramolecule | Name: RodA-PBP2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Nanodisc were formed using MSP1E3D1 and POPG lipid |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 111.803 KDa |
-Macromolecule #1: Peptidoglycan glycosyltransferase MrdB
| Macromolecule | Name: Peptidoglycan glycosyltransferase MrdB / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 40.508766 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MTDNPNKKTF WDKVHLDPTM LLILLALLVY SALVIWSASG QDIGMMERKI GQIAMGLVIM VVMAQIPPRV YEGWAPYLYI ICIILLVAV DAFGAISKGA QRWLDLGIVR FQPSEIAKIA VPLMVARFIN RDVCPPSLKN TGIALVLIFM PTLLVAAQPD L GTSILVAL ...String: MTDNPNKKTF WDKVHLDPTM LLILLALLVY SALVIWSASG QDIGMMERKI GQIAMGLVIM VVMAQIPPRV YEGWAPYLYI ICIILLVAV DAFGAISKGA QRWLDLGIVR FQPSEIAKIA VPLMVARFIN RDVCPPSLKN TGIALVLIFM PTLLVAAQPD L GTSILVAL SGLFVLFLSG LSWRLIGVAV VLVAAFIPIL WFFLMHDYQR QRVMMLLDPE SDPLGAGYHI IQSKIAIGSG GL RGKGWLH GTQSQLEFLP ERHTDFIFAV LAEELGLVGI LILLALYILL IMRGLWIAAR AQTTFGRVMA GGLMLILFVY VFV NIGMVS GILPVVGVPL PLVSYGGSAL IVLMAGFGIV MSIHTHRKML SKSV UniProtKB: Peptidoglycan glycosyltransferase MrdB |
-Macromolecule #2: Peptidoglycan D,D-transpeptidase MrdA
| Macromolecule | Name: Peptidoglycan D,D-transpeptidase MrdA / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 70.943414 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MKLQNSFRDY TAESALFVRR ALVAFLGILL LTGVLIANLY NLQIVRFTDY QTRSNENRIK LVPIAPSRGI IYDRNGIPLA LNRTIYQIE MMPEKVDNVQ QTLDALRSVV DLTDDDIAAF RKERARSHRF TSIPVKTNLT EVQVARFAVN QYRFPGVEVK G YKRRYYPY ...String: MKLQNSFRDY TAESALFVRR ALVAFLGILL LTGVLIANLY NLQIVRFTDY QTRSNENRIK LVPIAPSRGI IYDRNGIPLA LNRTIYQIE MMPEKVDNVQ QTLDALRSVV DLTDDDIAAF RKERARSHRF TSIPVKTNLT EVQVARFAVN QYRFPGVEVK G YKRRYYPY GSALTHVIGY VSKINDKDVE RLNNDGKLAN YAATHDIGKL GIERYYEDVL HGQTGYEEVE VNNRGRVIRQ LK EVPPQAG HDIYLTLDLK LQQYIETLLA GSRAAVVVTD PRTGGVLALV STPSYDPNLF VDGISSKDYS ALLNDPNTPL VNR ATQGVY PPASTVKPYV AVSALSAGVI TRNTTLFDPG WWQLPGSEKR YRDWKKWGHG RLNVTRSLEE SADTFFYQVA YDMG IDRLS EWMGKFGYGH YTGIDLAEER SGNMPTREWK QKRFKKPWYQ GDTIPVGIGQ GYWTATPIQM SKALMILIND GIVKV PHLL MSTAEDGKQV PWVQPHEPPV GDIHSGYWEL AKDGMYGVAN RPNGTAHKYF ASAPYKIAAK SGTAQVFGLK ANETYN AHK IAERLRDHKL MTAFAPYNNP QVAVAMILEN GGAGPAVGTL MRQILDHIML GDNNTDLPAE NPAVAAAEDH UniProtKB: Peptidoglycan D,D-transpeptidase MrdA |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.66 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 11120 / Average exposure time: 2.5 sec. / Average electron dose: 58.5 e/Å2 |
- Image processing
Image processing
| Particle selection | Number selected: 4415933 |
|---|---|
| Startup model | Type of model: OTHER |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2.12 and 3.2) / Software - details: Local refinement / Number images used: 399000 |
 Movie
Movie Controller
Controller