+ Open data
Open data
- Basic information
Basic information
| Entry | 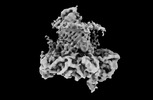 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of the EcBAM/EspP(beta8-12) complex | ||||||||||||
 Map data Map data | The cryo-EM structure of the EcBAM/EspP(beta8-12) complex | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  membrane proteins / membrane proteins /  protein folding / protein folding /  BAM complex / BAM complex /  outer membrane protein / outer membrane protein /  MEMBRANE PROTEIN MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationBam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / protein insertion into membrane / carbohydrate transmembrane transporter activity /  Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / cell outer membrane / outer membrane-bounded periplasmic space / Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / cell outer membrane / outer membrane-bounded periplasmic space /  periplasmic space / serine-type endopeptidase activity / periplasmic space / serine-type endopeptidase activity /  cell surface ...Bam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / protein insertion into membrane / carbohydrate transmembrane transporter activity / cell surface ...Bam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / protein insertion into membrane / carbohydrate transmembrane transporter activity /  Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / cell outer membrane / outer membrane-bounded periplasmic space / Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / cell outer membrane / outer membrane-bounded periplasmic space /  periplasmic space / serine-type endopeptidase activity / periplasmic space / serine-type endopeptidase activity /  cell surface / cell surface /  proteolysis / extracellular region / proteolysis / extracellular region /  membrane / identical protein binding membrane / identical protein bindingSimilarity search - Function | ||||||||||||
| Biological species |   Escherichia coli (E. coli) / Escherichia coli (E. coli) /   Escherichia coli O157 (bacteria) Escherichia coli O157 (bacteria) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.2 Å cryo EM / Resolution: 4.2 Å | ||||||||||||
 Authors Authors | Wu R / Noinaj N | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: BAM orchestrates OMP biogenesis using a beta-templating mechanism Authors: Wu R / Ryoo D / Kuo KM / Gumbart JC / Noinaj N | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40700.map.gz emd_40700.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40700-v30.xml emd-40700-v30.xml emd-40700.xml emd-40700.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40700.png emd_40700.png | 43.2 KB | ||
| Filedesc metadata |  emd-40700.cif.gz emd-40700.cif.gz | 6.9 KB | ||
| Others |  emd_40700_half_map_1.map.gz emd_40700_half_map_1.map.gz emd_40700_half_map_2.map.gz emd_40700_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40700 http://ftp.pdbj.org/pub/emdb/structures/EMD-40700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40700 | HTTPS FTP |
-Related structure data
| Related structure data |  8sqaMC  8sprC  8sqbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40700.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40700.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The cryo-EM structure of the EcBAM/EspP(beta8-12) complex | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_40700_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_40700_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BAM/EspP(beta1-12)
| Entire | Name: BAM/EspP(beta1-12) |
|---|---|
| Components |
|
-Supramolecule #1: BAM/EspP(beta1-12)
| Supramolecule | Name: BAM/EspP(beta1-12) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: Outer membrane protein assembly factor BamA
| Macromolecule | Name: Outer membrane protein assembly factor BamA / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 88.100305 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: VVKDIHFEGL QRVAVGAALL SMPVRTGDTV NDEDISNTIR ALFATGNFED VRVLRDGDTL LVQVKERPTI ASITFSGNKS VKDDMLKQN LEASGVRVGE SLDRTTIADI EKGLEDFYYS VGKYSASVKA VVTPLPRNRV DLKLVFQEGV SAEIQQINIV G NHAFTTDE ...String: VVKDIHFEGL QRVAVGAALL SMPVRTGDTV NDEDISNTIR ALFATGNFED VRVLRDGDTL LVQVKERPTI ASITFSGNKS VKDDMLKQN LEASGVRVGE SLDRTTIADI EKGLEDFYYS VGKYSASVKA VVTPLPRNRV DLKLVFQEGV SAEIQQINIV G NHAFTTDE LISHFQLRDE VPWWNVVGDR KYQKQKLAGD LETLRSYYLD RGYARFNIDS TQVSLTPDKK GIYVTVNITE GD QYKLSGV EVSGNLAGHS AEIEQLTKIE PGELYNGTKV TKMEDDIKKL LGRYGYAYPR VQSMPEINDA DKTVKLRVNV DAG NRFYVR KIRFEGNDTS KDAVLRREMR QMEGAWLGSD LVDQGKERLN RLGFFETVDT DTQRVPGSPD QVDVVYKVKE RNTG SFNFG IGYGTESGVS FQAGVQQDNW LGTGYAVGIN GTKNDYQTYA ELSVTNPYFT VDGVSLGGRL FYNDFQADDA DLSDY TNKS YGTDVTLGFP INEYNSLRAG LGYVHNSLSN MQPQVAMWRY LYSMGEHPST SDQDNSFKTD DFTFNYGWTY NKLDRG YFP TDGSRVNLTG KVTIPGSDNE YYKVTLDTAT YVPIDDDHKW VVLGRTRWGY GDGLGGKEMP FYENFYAGGS STVRGFQ SN TIGPKAVYFP HQASNYDPDY DYECATQDGA KDLCKSDDAV GGNAMAVASL EFITPTPFIS DKYANSVRTS FFWDMGTV W DTNWDSSQYS GYPDYSDPSN IRMSAGIALQ WMSPLGPLVF SYAQPFKKYD GDKAEQFQFN CGKTW UniProtKB: Outer membrane protein assembly factor BamA |
-Macromolecule #2: Outer membrane protein assembly factor BamB
| Macromolecule | Name: Outer membrane protein assembly factor BamB / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 39.77923 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: SLFNSEEDVV KMSPLPTVEN QFTPTTAWST SVGSGIGNFY SNLHPALADN VVYAADRAGL VKALNADDGK EIWSVSLAEK DGWFSKEPA LLSGGVTVSG GHVYIGSEKA QVYALNTSDG TVAWQTKVAG EALSRPVVSD GLVLIHTSNG QLQALNEADG A VKWTVNLD ...String: SLFNSEEDVV KMSPLPTVEN QFTPTTAWST SVGSGIGNFY SNLHPALADN VVYAADRAGL VKALNADDGK EIWSVSLAEK DGWFSKEPA LLSGGVTVSG GHVYIGSEKA QVYALNTSDG TVAWQTKVAG EALSRPVVSD GLVLIHTSNG QLQALNEADG A VKWTVNLD MPSLSLRGES APTTAFGAAV VGGDNGRVSA VLMEQGQMIW QQRISQATGS TEIDRLSDVD TTPVVVNGVV FA LAYNGNL TALDLRSGQI MWKRELGSVN DFIVDGNRIY LVDQNDRVMA LTIDGGVTLW TQSDLLHRLL TSPVLYNGNL VVG DSEGYL HWINVEDGRF VAQQKVDSSG FQTEPVAADG KLLIQAKDGT VYSITR UniProtKB: Outer membrane protein assembly factor BamB |
-Macromolecule #3: Outer membrane protein assembly factor BamC
| Macromolecule | Name: Outer membrane protein assembly factor BamC / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 34.298109 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: SSDSRYKRQV SGDEAYLEAA PLAELHAPAG MILPVTSGDY AIPVTNGSGA VGKALDIRPP AQPLALVSGA RTQFTGDTAS LLVENGRGN TLWPQVVSVL QAKNYTITQR DDAGQTLTTD WVQWNRLDED EQYRGRYQIS VKPQGYQQAV TVKLLNLEQA G KPVADAAS ...String: SSDSRYKRQV SGDEAYLEAA PLAELHAPAG MILPVTSGDY AIPVTNGSGA VGKALDIRPP AQPLALVSGA RTQFTGDTAS LLVENGRGN TLWPQVVSVL QAKNYTITQR DDAGQTLTTD WVQWNRLDED EQYRGRYQIS VKPQGYQQAV TVKLLNLEQA G KPVADAAS MQRYSTEMMN VISAGLDKSA TDAANAAQNR ASTTMDVQSA ADDTGLPMLV VRGPFNVVWQ RLPAALEKVG MK VTDSTRS QGNMAVTYKP LSDSDWQELG ASDPGLASGD YKLQVGDLDN RSSLQFIDPK GHTLTQSQND ALVAVFQAAF SK UniProtKB: Outer membrane protein assembly factor BamC |
-Macromolecule #4: Outer membrane protein assembly factor BamD
| Macromolecule | Name: Outer membrane protein assembly factor BamD / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 25.713676 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: SGSKEEVPDN PPNEIYATAQ QKLQDGNWRQ AITQLEALDN RYPFGPYSQQ VQLDLIYAYY KNADLPLAQA AIDRFIRLNP THPNIDYVM YMRGLTNMAL DDSALQGFFG VDRSDRDPQH ARAAFSDFSK LVRGYPNSQY TTDATKRLVF LKDRLAKYEY S VAEYYTER ...String: SGSKEEVPDN PPNEIYATAQ QKLQDGNWRQ AITQLEALDN RYPFGPYSQQ VQLDLIYAYY KNADLPLAQA AIDRFIRLNP THPNIDYVM YMRGLTNMAL DDSALQGFFG VDRSDRDPQH ARAAFSDFSK LVRGYPNSQY TTDATKRLVF LKDRLAKYEY S VAEYYTER GAWVAVVNRV EGMLRDYPDT QATRDALPLM ENAYRQMQMN AQAEKVAKII AANSSNT UniProtKB: Outer membrane protein assembly factor BamD |
-Macromolecule #5: Outer membrane protein assembly factor BamE
| Macromolecule | Name: Outer membrane protein assembly factor BamE / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 10.28841 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: STLERVVYRP DINQGNYLTA NDVSKIRVGM TQQQVAYALG TPLMSDPFGT NTWFYVFRQQ PGHEGVTQQT LTLTFNSSGV LTNIDNKPA LSGN UniProtKB: Outer membrane protein assembly factor BamE |
-Macromolecule #6: Maltose/maltodextrin-binding periplasmic protein,Autotransporter ...
| Macromolecule | Name: Maltose/maltodextrin-binding periplasmic protein,Autotransporter protein EspP translocator type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli O157 (bacteria) Escherichia coli O157 (bacteria) |
| Molecular weight | Theoretical: 60.668305 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MGAKIEEGKL VIWINGDKGY NGLAEVGKKF EKDTGIKVTV EHPDKLEEKF PQVAATGDGP DIIFWAHDRF GGYAQSGLLA EITPDKAFQ DKLYPFTWDA VRYNGKLIAY PIAVEALSLI YNKDLLPNPP KTWEEIPALD KELKAKGKSA LMFNLQEPYF T WPLIAADG ...String: MGAKIEEGKL VIWINGDKGY NGLAEVGKKF EKDTGIKVTV EHPDKLEEKF PQVAATGDGP DIIFWAHDRF GGYAQSGLLA EITPDKAFQ DKLYPFTWDA VRYNGKLIAY PIAVEALSLI YNKDLLPNPP KTWEEIPALD KELKAKGKSA LMFNLQEPYF T WPLIAADG GYAFKYENGK YDIKDVGVDN AGAKAGLTFL VDLIKNKHMN ADTDYSIAEA AFNKGETAMT INGPWAWSNI DT SKVNYGV TVLPTFKGQP SKPFVGVLSA GINAASPNKE LAKEFLENYL LTDEGLEAVN KDKPLGAVAL KSYEEELVKD PRI AATMEN AQKGEIMPNI PQMSAFWYAV RTAVINAASG RQTVDEALKD AQTGSIELVS APKDTNENVF KASKQTIGFS DVTP VITTR ETDDKITWSL TGYNTVANKE ATRNAAALFS VDYKAFLNEV KDKDYNPLIG RTGCDVGKSF SGKDWKVTAR AGLGY QFDL LANGETVLRD ASGEKRIKGE KDSRMLMSVG LNAEIRDNVR FGLEFEKSAF GKYNVDNAVN ANFRYSF UniProtKB: Maltose/maltodextrin-binding periplasmic protein, Serine protease EspP |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 53.47 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 92335 |
 Movie
Movie Controller
Controller



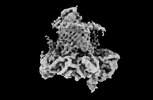
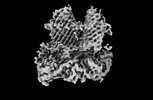
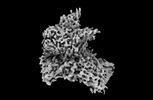








 Z
Z Y
Y X
X