[English] 日本語
 Yorodumi
Yorodumi- EMDB-38244: Open state of central tail fiber of bacteriophage lambda upon bin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 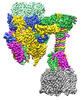 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
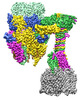
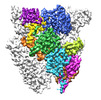
| Title | Open state of central tail fiber of bacteriophage lambda upon binding to LamB | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Bacteriophage / Bacteriophage /  caudovirales / caudovirales /  siphoviridae / siphoviridae /  phage lambda / host recognition / LamB / phage lambda / host recognition / LamB /  cryo-EM / cryo-EM /  VIRUS VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont genome ejection through host cell envelope, long flexible tail mechanism / viral tail assembly / virus tail / host cell cytoplasm / entry receptor-mediated virion attachment to host cell / receptor-mediated virion attachment to host cell / virion attachment to host cell Similarity search - Function | |||||||||
| Biological species |   Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.57 Å cryo EM / Resolution: 3.57 Å | |||||||||
 Authors Authors | Ge XF / Wang JW | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of lambda tail with LamB Authors: Ge XF / Wang JW | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38244.map.gz emd_38244.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38244-v30.xml emd-38244-v30.xml emd-38244.xml emd-38244.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38244_fsc.xml emd_38244_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_38244.png emd_38244.png | 172.5 KB | ||
| Filedesc metadata |  emd-38244.cif.gz emd-38244.cif.gz | 5.7 KB | ||
| Others |  emd_38244_half_map_1.map.gz emd_38244_half_map_1.map.gz emd_38244_half_map_2.map.gz emd_38244_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38244 http://ftp.pdbj.org/pub/emdb/structures/EMD-38244 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38244 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38244 | HTTPS FTP |
-Related structure data
| Related structure data |  8xciMC  8xcgC  8xcjC  8xckC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38244.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38244.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.0742 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38244_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
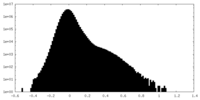
| Density Histograms |
-Half map: #1
| File | emd_38244_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Bacteriophage lambda tail with LamB
| Entire | Name: Bacteriophage lambda tail with LamB |
|---|---|
| Components |
|
-Supramolecule #1: Bacteriophage lambda tail with LamB
| Supramolecule | Name: Bacteriophage lambda tail with LamB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
-Macromolecule #1: Tip attachment protein J
| Macromolecule | Name: Tip attachment protein J / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 124.550625 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MGKGSSKGHT PREAKDNLKS TQLLSVIDAI SEGPIEGPVD GLKSVLLNST PVLDTEGNTN ISGVTVVFRA GEQEQTPPEG FESSGSETV LGTEVKYDTP ITRTITSANI DRLRFTFGVQ ALVETTSKGD RNPSEVRLLV QIQRNGGWVT EKDITIKGKT T SQYLASVV ...String: MGKGSSKGHT PREAKDNLKS TQLLSVIDAI SEGPIEGPVD GLKSVLLNST PVLDTEGNTN ISGVTVVFRA GEQEQTPPEG FESSGSETV LGTEVKYDTP ITRTITSANI DRLRFTFGVQ ALVETTSKGD RNPSEVRLLV QIQRNGGWVT EKDITIKGKT T SQYLASVV MGNLPPRPFN IRMRRMTPDS TTDQLQNKTL WSSYTEIIDV KQCYPNTALV GVQVDSEQFG SQQVSRNYHL RG RILQVPS NYNPQTRQYS GIWDGTFKPA YSNNMAWCLW DMLTHPRYGM GKRLGAADVD KWALYVIGQY CDQSVPDGFG GTE PRITCN AYLTTQRKAW DVLSDFCSAM RCMPVWNGQT LTFVQDRPSD KTWTYNRSNV VMPDDGAPFR YSFSALKDRH NAVE VNWID PNNGWETATE LVEDTQAIAR YGRNVTKMDA FGCTSRGQAH RAGLWLIKTE LLETQTVDFS VGAEGLRHVP GDVIE ICDD DYAGISTGGR VLAVNSQTRT LTLDREITLP SSGTALISLV DGSGNPVSVE VQSVTDGVKV KVSRVPDGVA EYSVWE LKL PTLRQRLFRC VSIRENDDGT YAITAVQHVP EKEAIVDNGA HFDGEQSGTV NGVTPPAVQH LTAEVTADSG EYQVLAR WD TPKVVKGVSF LLRLTVTADD GSERLVSTAR TTETTYRFTQ LALGNYRLTV RAVNAWGQQG DPASVSFRIA APAAPSRI E LTPGYFQITA TPHLAVYDPT VQFEFWFSEK QIADIRQVET STRYLGTALY WIAASINIKP GHDYYFYIRS VNTVGKSAF VEAVGRASDD AEGYLDFFKG KITESHLGKE LLEKVELTED NASRLEEFSK EWKDASDKWN AMWAVKIEQT KDGKHYVAGI GLSMEDTEE GKLSQFLVAA NRIAFIDPAN GNETPMFVAQ GNQIFMNDVF LKRLTAPTIT SGGNPPAFSL TPDGKLTAKN A DISGSVNA NSGTLSNVTI AENCTINGTL RAEKIVGDIV KAASAAFPRQ RESSVDWPSG TRTVTVTDDH PFDRQIVVLP LT FRGSKRT VSGRTTYSMC YLKVLMNGAV IYDGAANEAV QVFSRIVDMP AGRGNVILTF TLTSTRHSAD IPPYTFASDV QVM VIKKQA LGISVV UniProtKB: Tip attachment protein J |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X