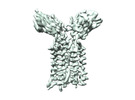
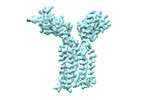
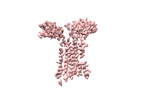
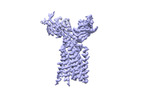
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
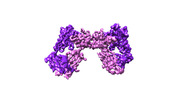
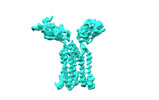
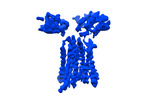
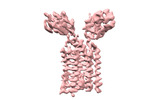
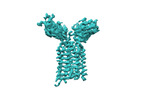
| Title | T cell receptor delta 2 gamma 9 with F283A, F290A, and F291A | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  immune / immune /  receptor / receptor /  IMMUNE SYSTEM IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationMHC protein binding / regulation of lymphocyte apoptotic process / gamma-delta T cell receptor complex /  Fc-gamma receptor III complex / T cell anergy / positive regulation of T cell anergy / positive regulation of cell-cell adhesion mediated by integrin / Fc-gamma receptor signaling pathway / CD4-positive, alpha-beta T cell proliferation / gamma-delta T cell activation ...MHC protein binding / regulation of lymphocyte apoptotic process / gamma-delta T cell receptor complex / Fc-gamma receptor III complex / T cell anergy / positive regulation of T cell anergy / positive regulation of cell-cell adhesion mediated by integrin / Fc-gamma receptor signaling pathway / CD4-positive, alpha-beta T cell proliferation / gamma-delta T cell activation ...MHC protein binding / regulation of lymphocyte apoptotic process / gamma-delta T cell receptor complex /  Fc-gamma receptor III complex / T cell anergy / positive regulation of T cell anergy / positive regulation of cell-cell adhesion mediated by integrin / Fc-gamma receptor signaling pathway / CD4-positive, alpha-beta T cell proliferation / gamma-delta T cell activation / negative thymic T cell selection / positive regulation of CD4-positive, alpha-beta T cell proliferation / positive regulation of protein localization to cell surface / alpha-beta T cell receptor complex / positive thymic T cell selection / signal complex assembly / Nef and signal transduction / positive regulation of cell-matrix adhesion / Fc-gamma receptor III complex / T cell anergy / positive regulation of T cell anergy / positive regulation of cell-cell adhesion mediated by integrin / Fc-gamma receptor signaling pathway / CD4-positive, alpha-beta T cell proliferation / gamma-delta T cell activation / negative thymic T cell selection / positive regulation of CD4-positive, alpha-beta T cell proliferation / positive regulation of protein localization to cell surface / alpha-beta T cell receptor complex / positive thymic T cell selection / signal complex assembly / Nef and signal transduction / positive regulation of cell-matrix adhesion /  T cell receptor complex / smoothened signaling pathway / establishment or maintenance of cell polarity / Translocation of ZAP-70 to Immunological synapse / Phosphorylation of CD3 and TCR zeta chains / dendrite development / T cell receptor complex / smoothened signaling pathway / establishment or maintenance of cell polarity / Translocation of ZAP-70 to Immunological synapse / Phosphorylation of CD3 and TCR zeta chains / dendrite development /  small molecule binding / Generation of second messenger molecules / alpha-beta T cell activation / small molecule binding / Generation of second messenger molecules / alpha-beta T cell activation /  immunological synapse / FCGR activation / PD-1 signaling / positive regulation of interleukin-4 production / Role of phospholipids in phagocytosis / negative regulation of smoothened signaling pathway / positive regulation of calcium-mediated signaling / positive regulation of T cell proliferation / immunological synapse / FCGR activation / PD-1 signaling / positive regulation of interleukin-4 production / Role of phospholipids in phagocytosis / negative regulation of smoothened signaling pathway / positive regulation of calcium-mediated signaling / positive regulation of T cell proliferation /  protein tyrosine kinase binding / T cell costimulation / positive regulation of interleukin-2 production / protein tyrosine kinase binding / T cell costimulation / positive regulation of interleukin-2 production /  T cell receptor binding / cerebellum development / T cell receptor binding / cerebellum development /  T cell activation / FCGR3A-mediated IL10 synthesis / calcium-mediated signaling / apoptotic signaling pathway / FCGR3A-mediated phagocytosis / clathrin-coated endocytic vesicle membrane / Regulation of actin dynamics for phagocytic cup formation / T cell activation / FCGR3A-mediated IL10 synthesis / calcium-mediated signaling / apoptotic signaling pathway / FCGR3A-mediated phagocytosis / clathrin-coated endocytic vesicle membrane / Regulation of actin dynamics for phagocytic cup formation /  SH3 domain binding / peptide antigen binding / SH3 domain binding / peptide antigen binding /  cell surface receptor protein tyrosine kinase signaling pathway / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / positive regulation of peptidyl-tyrosine phosphorylation / cell-cell junction / positive regulation of type II interferon production / transmembrane signaling receptor activity / signaling receptor complex adaptor activity / Cargo recognition for clathrin-mediated endocytosis / cell surface receptor protein tyrosine kinase signaling pathway / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / positive regulation of peptidyl-tyrosine phosphorylation / cell-cell junction / positive regulation of type II interferon production / transmembrane signaling receptor activity / signaling receptor complex adaptor activity / Cargo recognition for clathrin-mediated endocytosis /  protein transport / Downstream TCR signaling / protein complex oligomerization / protein transport / Downstream TCR signaling / protein complex oligomerization /  Clathrin-mediated endocytosis / Clathrin-mediated endocytosis /  cell body / T cell receptor signaling pathway / protein-containing complex assembly / regulation of apoptotic process / cell body / T cell receptor signaling pathway / protein-containing complex assembly / regulation of apoptotic process /  dendritic spine / dendritic spine /  adaptive immune response / cell surface receptor signaling pathway / G protein-coupled receptor signaling pathway / protein heterodimerization activity / external side of plasma membrane / negative regulation of gene expression / adaptive immune response / cell surface receptor signaling pathway / G protein-coupled receptor signaling pathway / protein heterodimerization activity / external side of plasma membrane / negative regulation of gene expression /  innate immune response / positive regulation of gene expression / innate immune response / positive regulation of gene expression /  protein kinase binding / protein kinase binding /  Golgi apparatus / Golgi apparatus /  endoplasmic reticulum / protein homodimerization activity / identical protein binding / endoplasmic reticulum / protein homodimerization activity / identical protein binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.8 Å cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Xin W / Huang B / Chi X / Liu Y / Xu M / Zhang Y / Li X / Su Q / Zhou Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structures of human γδ T cell receptor-CD3 complex. Authors: Weizhi Xin / Bangdong Huang / Ximin Chi / Yuehua Liu / Mengjiao Xu / Yuanyuan Zhang / Xu Li / Qiang Su / Qiang Zhou /  Abstract: Gamma delta (γδ) T cells, a unique T cell subgroup, are crucial in various immune responses and immunopathology. The γδ T cell receptor (TCR), generated by γδ T cells, recognizes a diverse ...Gamma delta (γδ) T cells, a unique T cell subgroup, are crucial in various immune responses and immunopathology. The γδ T cell receptor (TCR), generated by γδ T cells, recognizes a diverse range of antigens independently of the major histocompatibility complex. The γδ TCR associates with CD3 subunits, initiating T cell activation and holding great potential in immunotherapy. Here, we report the structures of two prototypical human Vγ9Vδ2 and Vγ5Vδ1 TCR-CD3 complexes, unveiling two distinct assembly mechanisms that depend on Vγ usage. The Vγ9Vδ2 TCR-CD3 complex is monomeric, with considerable conformational flexibility in the TCRγ/TCRδ extracellular domain (ECD) and connecting peptides (CPs). The length of CPs regulates the ligand association and T cell activation. Additionally, a cholesterol-like molecule wedges into the transmembrane region, exerting an inhibitory role in TCR signaling. The Vγ5Vδ1 TCR-CD3 complex displays a dimeric architecture, where two protomers nestle back-to-back via their Vγ5 domains of TCR ECDs. Our biochemical and biophysical assays further corroborate the dimeric structure. Importantly, the dimeric form of the Vγ5Vδ1 TCR is essential for T cell activation. These findings reveal organizing principles of the γδ TCR-CD3 complex, providing insights into the γδ TCR unique properties and facilitating immunotherapeutic interventions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37914.map.gz emd_37914.map.gz | 27.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37914-v30.xml emd-37914-v30.xml emd-37914.xml emd-37914.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37914.png emd_37914.png | 50.4 KB | ||
| Filedesc metadata |  emd-37914.cif.gz emd-37914.cif.gz | 6.5 KB | ||
| Others |  emd_37914_half_map_1.map.gz emd_37914_half_map_1.map.gz emd_37914_half_map_2.map.gz emd_37914_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37914 http://ftp.pdbj.org/pub/emdb/structures/EMD-37914 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37914 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37914 | HTTPS FTP |
-Related structure data
| Related structure data |  8wy0MC  8jbvC  8jc0C  8jcbC  8wxeC  8wyiC  8yc0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37914.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37914.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.0773 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37914_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37914_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : T cell receptor delta 2 gamma 9 with F283A, F290A, F291A
| Entire | Name: T cell receptor delta 2 gamma 9 with F283A, F290A, F291A |
|---|---|
| Components |
|
-Supramolecule #1: T cell receptor delta 2 gamma 9 with F283A, F290A, F291A
| Supramolecule | Name: T cell receptor delta 2 gamma 9 with F283A, F290A, F291A type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: T-cell surface glycoprotein CD3 zeta chain
| Macromolecule | Name: T-cell surface glycoprotein CD3 zeta chain / type: protein_or_peptide / ID: 1 / Details: 168-195 Twin-Strep tag / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 21.809695 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKWKALFTAA ILQAQLPITE AQSFGLLDPK LCYLLDGILF IYGVILTALF LRVKFSRSAD APAYQQGQNQ LYNELNLGRR EEYDVLDKR RGRDPEMGGK PQRRKNPQEG LYNELQKDKM AEAYSEIGMK GERRRGKGHD GLYQGLSTAT KDTYDALHMQ A LPPRAAAW ...String: MKWKALFTAA ILQAQLPITE AQSFGLLDPK LCYLLDGILF IYGVILTALF LRVKFSRSAD APAYQQGQNQ LYNELNLGRR EEYDVLDKR RGRDPEMGGK PQRRKNPQEG LYNELQKDKM AEAYSEIGMK GERRRGKGHD GLYQGLSTAT KDTYDALHMQ A LPPRAAAW SHPQFEKGGG SGGGSGGSAW SHPQFEK UniProtKB:  T-cell surface glycoprotein CD3 zeta chain T-cell surface glycoprotein CD3 zeta chain |
-Macromolecule #2: T-cell surface glycoprotein CD3 delta chain
| Macromolecule | Name: T-cell surface glycoprotein CD3 delta chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.949537 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEHSTFLSGL VLATLLSQVS PFKIPIEELE DRVFVNCNTS ITWVEGTVGT LLSDITRLDL GKRILDPRGI YRCNGTDIYK DKESTVQVH YRMCQSCVEL DPATVAGIIV TDVIATLLLA LGVFCFAGHE TGRLSGAADT QALLRNDQVY QPLRDRDDAQ Y SHLGGNWA RNK UniProtKB: T-cell surface glycoprotein CD3 delta chain |
-Macromolecule #3: T-cell surface glycoprotein CD3 epsilon chain
| Macromolecule | Name: T-cell surface glycoprotein CD3 epsilon chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.174227 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQSGTHWRVL GLCLLSVGVW GQDGNEEMGG ITQTPYKVSI SGTTVILTCP QYPGSEILWQ HNDKNIGGDE DDKNIGSDED HLSLKEFSE LEQSGYYVCY PRGSKPEDAN FYLYLRARVC ENCMEMDVMS VATIVIVDIC ITGGLLLLVY YWSKNRKAKA K PVTRGAGA ...String: MQSGTHWRVL GLCLLSVGVW GQDGNEEMGG ITQTPYKVSI SGTTVILTCP QYPGSEILWQ HNDKNIGGDE DDKNIGSDED HLSLKEFSE LEQSGYYVCY PRGSKPEDAN FYLYLRARVC ENCMEMDVMS VATIVIVDIC ITGGLLLLVY YWSKNRKAKA K PVTRGAGA GGRQRGQNKE RPPPVPNPDY EPIRKGQRDL YSGLNQRRI UniProtKB: T-cell surface glycoprotein CD3 epsilon chain |
-Macromolecule #4: T-cell surface glycoprotein CD3 gamma chain
| Macromolecule | Name: T-cell surface glycoprotein CD3 gamma chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.493457 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEQGKGLAVL ILAIILLQGT LAQSIKGNHL VKVYDYQEDG SVLLTCDAEA KNITWFKDGK MIGFLTEDKK KWNLGSNAKD PRGMYQCKG SQNKSKPLQV YYRMCQNCIE LNAATISGFL FAEIVSIFVL AVGVYFIAGQ DGVRQSRASD KQTLLPNDQL Y QPLKDRED DQYSHLQGNQ LRRN UniProtKB: T-cell surface glycoprotein CD3 gamma chain |
-Macromolecule #5: Signal peptide,flag tag,T cell receptor delta variable 2,T cell r...
| Macromolecule | Name: Signal peptide,flag tag,T cell receptor delta variable 2,T cell receptor delta constant type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.331457 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDMRVPAQLL GLLLLWLSGA RCMDYKDDDD KGGSETGAIE LVPEHQTVPV SIGVPATLRC SMKGEAIGNY YINWYRKTQG NTMTFIYRE KDIYGPGFKD NFQGDIDIAK NLAVLKILAP SERDEGSYYC ACDTLGMGGE YTDKLIFGKG TRVTVEPRSQ P HTKPSVFV ...String: MDMRVPAQLL GLLLLWLSGA RCMDYKDDDD KGGSETGAIE LVPEHQTVPV SIGVPATLRC SMKGEAIGNY YINWYRKTQG NTMTFIYRE KDIYGPGFKD NFQGDIDIAK NLAVLKILAP SERDEGSYYC ACDTLGMGGE YTDKLIFGKG TRVTVEPRSQ P HTKPSVFV MKNGTNVACL VKEFYPKDIR INLVSSKKIT EFDPAIVISP SGKYNAVKLG KYEDSNSVTC SVQHDNKTVH ST DFEVKTD STDHVKPKET ENTKQPSKSC HKPKAIVHTE KVNMMSLTVL GLRMLFAKTV AVNALLTAKL AAL UniProtKB: T cell receptor delta variable 2, T cell receptor delta constant |
-Macromolecule #6: Signal peptide,flag tag,T cell receptor gamma variable 9,T cell r...
| Macromolecule | Name: Signal peptide,flag tag,T cell receptor gamma variable 9,T cell receptor gamma constant 1 type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.406973 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDMRVPAQLL GLLLLWLSGA RCMDYKDDDD KGGSETGAGH LEQPQISSTK TLSKTARLEC VVSGITISAT SVYWYRERPG EVIQFLVSI SYDGTVRKES GIPSGKFEVD RIPETSTSTL TIHNVEKQDI ATYYCALWEA QQELGKKIKV FGPGTKLIIT D KQLDADVS ...String: MDMRVPAQLL GLLLLWLSGA RCMDYKDDDD KGGSETGAGH LEQPQISSTK TLSKTARLEC VVSGITISAT SVYWYRERPG EVIQFLVSI SYDGTVRKES GIPSGKFEVD RIPETSTSTL TIHNVEKQDI ATYYCALWEA QQELGKKIKV FGPGTKLIIT D KQLDADVS PKPTIFLPSI AETKLQKAGT YLCLLEKFFP DVIKIHWQEK KSNTILGSQE GNTMKTNDTY MKFSWLTVPE KS LDKEHRC IVRHENNKNG VDQEIIFPPI KTDVITMDPK DNCSKDANDT LLLQLTNTSA YYMYLLLLLK SVVYFAIITC CLL RRTAFC CNGEKS UniProtKB: T cell receptor gamma variable 9, T cell receptor gamma constant 1 |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 294780 |
 Movie
Movie Controller
Controller


























 Z
Z Y
Y X
X

















