+ Open data
Open data
- Basic information
Basic information
| Entry | 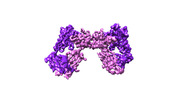 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
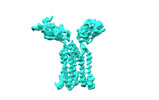
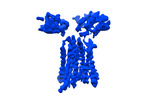
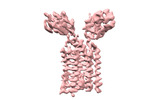
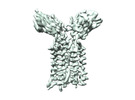
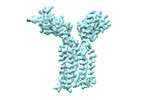
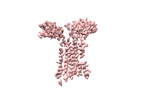
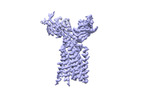
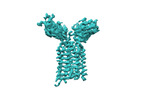
| Title | Extracellular domain of gamma delta TCR | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Receptor /  IMMUNE SYSTEM IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationgamma-delta T cell receptor complex / gamma-delta T cell activation /  T cell receptor complex / T cell receptor complex /  small molecule binding / T cell receptor signaling pathway / small molecule binding / T cell receptor signaling pathway /  adaptive immune response / adaptive immune response /  immune response / external side of plasma membrane / immune response / external side of plasma membrane /  innate immune response / innate immune response /  extracellular space / extracellular space /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
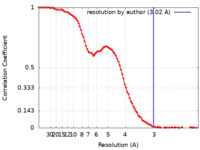
| Method |  single particle reconstruction / Resolution: 3.02 Å single particle reconstruction / Resolution: 3.02 Å | |||||||||
 Authors Authors | Xin W / Chi X / Huang B / Su Q / Zhou Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structures of human γδ T cell receptor-CD3 complex. Authors: Weizhi Xin / Bangdong Huang / Ximin Chi / Yuehua Liu / Mengjiao Xu / Yuanyuan Zhang / Xu Li / Qiang Su / Qiang Zhou /  Abstract: Gamma delta (γδ) T cells, a unique T cell subgroup, are crucial in various immune responses and immunopathology. The γδ T cell receptor (TCR), generated by γδ T cells, recognizes a diverse ...Gamma delta (γδ) T cells, a unique T cell subgroup, are crucial in various immune responses and immunopathology. The γδ T cell receptor (TCR), generated by γδ T cells, recognizes a diverse range of antigens independently of the major histocompatibility complex. The γδ TCR associates with CD3 subunits, initiating T cell activation and holding great potential in immunotherapy. Here, we report the structures of two prototypical human Vγ9Vδ2 and Vγ5Vδ1 TCR-CD3 complexes, unveiling two distinct assembly mechanisms that depend on Vγ usage. The Vγ9Vδ2 TCR-CD3 complex is monomeric, with considerable conformational flexibility in the TCRγ/TCRδ extracellular domain (ECD) and connecting peptides (CPs). The length of CPs regulates the ligand association and T cell activation. Additionally, a cholesterol-like molecule wedges into the transmembrane region, exerting an inhibitory role in TCR signaling. The Vγ5Vδ1 TCR-CD3 complex displays a dimeric architecture, where two protomers nestle back-to-back via their Vγ5 domains of TCR ECDs. Our biochemical and biophysical assays further corroborate the dimeric structure. Importantly, the dimeric form of the Vγ5Vδ1 TCR is essential for T cell activation. These findings reveal organizing principles of the γδ TCR-CD3 complex, providing insights into the γδ TCR unique properties and facilitating immunotherapeutic interventions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36147.map.gz emd_36147.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36147-v30.xml emd-36147-v30.xml emd-36147.xml emd-36147.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36147_fsc.xml emd_36147_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_36147.png emd_36147.png | 42.5 KB | ||
| Filedesc metadata |  emd-36147.cif.gz emd-36147.cif.gz | 5.4 KB | ||
| Others |  emd_36147_half_map_1.map.gz emd_36147_half_map_1.map.gz emd_36147_half_map_2.map.gz emd_36147_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36147 http://ftp.pdbj.org/pub/emdb/structures/EMD-36147 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36147 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36147 | HTTPS FTP |
-Related structure data
| Related structure data |  8jbvMC  8jc0C  8jcbC  8wxeC  8wy0C  8wyiC  8yc0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36147.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36147.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36147_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36147_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Extracellular domain of gamma delta TCR
| Entire | Name: Extracellular domain of gamma delta TCR |
|---|---|
| Components |
|
-Supramolecule #1: Extracellular domain of gamma delta TCR
| Supramolecule | Name: Extracellular domain of gamma delta TCR / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: T cell receptor delta variable 1,T cell receptor delta constant
| Macromolecule | Name: T cell receptor delta variable 1,T cell receptor delta constant type: protein_or_peptide / ID: 1 Details: Author stated: 1-22:Signal peptide, 23-31:Flag tag. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.320574 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDMRVPAQLL GLLLLWLSGA RCMDYKDDDD KGGSETGAQK VTQAQSSVSM PVRKAVTLNC LYETSWWSYY IFWYKQLPSK EMIFLIRQG SDEQNAKSGR YSVNFKKAAK SVALTISALQ LEDSAKYFCA LGDPGGLNTD KLIFGKGTRV TVEPRSQPHT K PSVFVMKN ...String: MDMRVPAQLL GLLLLWLSGA RCMDYKDDDD KGGSETGAQK VTQAQSSVSM PVRKAVTLNC LYETSWWSYY IFWYKQLPSK EMIFLIRQG SDEQNAKSGR YSVNFKKAAK SVALTISALQ LEDSAKYFCA LGDPGGLNTD KLIFGKGTRV TVEPRSQPHT K PSVFVMKN GTNVACLVKE FYPKDIRINL VSSKKITEFD PAIVISPSGK YNAVKLGKYE DSNSVTCSVQ HDNKTVHSTD FE VKTDSTD HVKPKETENT KQPSKSCHKP KAIVHTEKVN MMSLTVLGLR MLFAKTVAVN FLLTAKLFFL UniProtKB: T cell receptor delta variable 1, T cell receptor delta constant |
-Macromolecule #2: T cell receptor gamma variable 5,T cell receptor gamma constant 1
| Macromolecule | Name: T cell receptor gamma variable 5,T cell receptor gamma constant 1 type: protein_or_peptide / ID: 2 Details: Author stated: 1-22:Signal peptide, 23-31:Flag tag. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.755172 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDMRVPAQLL GLLLLWLSGA RCMDYKDDDD KGGSETGSSN LEGGTKSVTR PTRSSAEITC DLTVINAFYI HWYLHQEGKA PQRLLYYDV SNSKDVLESG LSPGKYYTHT PRRWSWILIL RNLIENDSGV YYCATWDRGN PKTHYYKKLF GSGTTLVVTD K QLDADVSP ...String: MDMRVPAQLL GLLLLWLSGA RCMDYKDDDD KGGSETGSSN LEGGTKSVTR PTRSSAEITC DLTVINAFYI HWYLHQEGKA PQRLLYYDV SNSKDVLESG LSPGKYYTHT PRRWSWILIL RNLIENDSGV YYCATWDRGN PKTHYYKKLF GSGTTLVVTD K QLDADVSP KPTIFLPSIA ETKLQKAGTY LCLLEKFFPD VIKIHWQEKK SNTILGSQEG NTMKTNDTYM KFSWLTVPEK SL DKEHRCI VRHENNKNGV DQEIIFPPIK TDVITMDPKD NCSKDANDTL LLQLTNTSAY YMYLLLLLKS VVYFAIITCC LLR RTAFCC NGEKS UniProtKB: T cell receptor gamma variable 5, T cell receptor gamma constant 1 |
-Experimental details
-Structure determination
 Processing Processing |  single particle reconstruction single particle reconstruction |
|---|---|
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X