[English] 日本語
 Yorodumi
Yorodumi- EMDB-36902: Cryo-EM structure of GSK256073 bound human hydroxy-carboxylic aci... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of GSK256073 bound human hydroxy-carboxylic acid receptor 2 (Local refinement) | |||||||||
 Map data Map data | sharpening map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  GPCR / GPCR /  G-Protein / G-Protein /  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  signaling signaling | |||||||||
| Function / homology |  Function and homology information Function and homology information nicotinic acid receptor activity / Hydroxycarboxylic acid-binding receptors / neutrophil apoptotic process / Class A/1 (Rhodopsin-like receptors) / positive regulation of neutrophil apoptotic process / positive regulation of adiponectin secretion / negative regulation of lipid catabolic process / nicotinic acid receptor activity / Hydroxycarboxylic acid-binding receptors / neutrophil apoptotic process / Class A/1 (Rhodopsin-like receptors) / positive regulation of neutrophil apoptotic process / positive regulation of adiponectin secretion / negative regulation of lipid catabolic process /  cell junction / G alpha (i) signalling events / G protein-coupled receptor signaling pathway / cell junction / G alpha (i) signalling events / G protein-coupled receptor signaling pathway /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
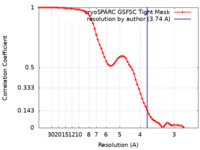
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.74 Å cryo EM / Resolution: 3.74 Å | |||||||||
 Authors Authors | Park JH / Ishimoto N / Park SY | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2 Authors: Park JH / Kawakami K / Ishimoto N / Ikuta T / Ohki M / Ekimoto T / Ikeguchi M / Lee DS / Lee YH / Tame JRH / Inoue A / Park SY | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36902.map.gz emd_36902.map.gz | 28.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36902-v30.xml emd-36902-v30.xml emd-36902.xml emd-36902.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36902_fsc.xml emd_36902_fsc.xml | 7 KB | Display |  FSC data file FSC data file |
| Images |  emd_36902.png emd_36902.png | 72.6 KB | ||
| Masks |  emd_36902_msk_1.map emd_36902_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36902.cif.gz emd-36902.cif.gz | 5.9 KB | ||
| Others |  emd_36902_additional_1.map.gz emd_36902_additional_1.map.gz emd_36902_half_map_1.map.gz emd_36902_half_map_1.map.gz emd_36902_half_map_2.map.gz emd_36902_half_map_2.map.gz | 15.2 MB 28.3 MB 28.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36902 http://ftp.pdbj.org/pub/emdb/structures/EMD-36902 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36902 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36902 | HTTPS FTP |
-Related structure data
| Related structure data |  8k5dMC  8k5bC  8k5cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36902.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36902.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpening map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.245 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36902_msk_1.map emd_36902_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
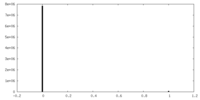
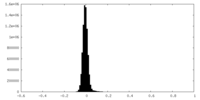
| Density Histograms |
-Additional map: map
| File | emd_36902_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||
| Projections & Slices |
| ||||||||||||
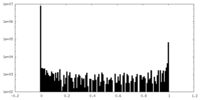
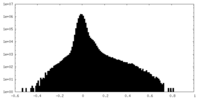
| Density Histograms |
-Half map: half map 2
| File | emd_36902_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
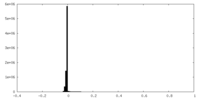
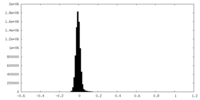
| Density Histograms |
-Half map: half map 1
| File | emd_36902_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
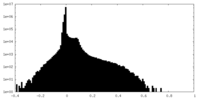
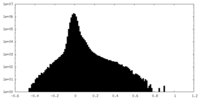
| Density Histograms |
- Sample components
Sample components
-Entire : GSK256073 bound human hydroxy-carboxylic acid receptor 2
| Entire | Name: GSK256073 bound human hydroxy-carboxylic acid receptor 2 |
|---|---|
| Components |
|
-Supramolecule #1: GSK256073 bound human hydroxy-carboxylic acid receptor 2
| Supramolecule | Name: GSK256073 bound human hydroxy-carboxylic acid receptor 2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Human hydroxycarboxylic acid receptor 2
| Macromolecule | Name: Human hydroxycarboxylic acid receptor 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.320297 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: GPGAPADLED NWETLNDNLK VIEKADNAAQ VKDALTKMRA AALDAQKATP PKLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKV KEAQAAAEQL KTTRNAYIQK YLEFMNRHHL QDHFLEIDKK NCCVFRDDFI VKVLPPVLGL EFIFGLLGNG L ALWIFCFH ...String: GPGAPADLED NWETLNDNLK VIEKADNAAQ VKDALTKMRA AALDAQKATP PKLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKV KEAQAAAEQL KTTRNAYIQK YLEFMNRHHL QDHFLEIDKK NCCVFRDDFI VKVLPPVLGL EFIFGLLGNG L ALWIFCFH LKSWKSSRIF LFNLAVADFL LIICLPFLMD NYVRRWDWKF GDIPCRLMLF MLAMNRQGSI IFLTVVAVDR YF RVVHPHH ALNKISNRTA AIISCLLWGI TIGLTVHLLK KKMPIQNGGA NLCSSFSICH TFQWHEAMFL LEFFLPLGII LFC SARIIW SLRQRQMDRH AKIKRAITFI MVVAIVFVIC FLPSVVVRIR IFWLLHTSGT QNCEVYRSVD LAFFITLSFT YMNS MLDPV VYYFSSPSFP NFFSTLINRC LQRKMTGEPD NNRSTSVELT GDPNKTRGAP EALMANSGEP WSPSYLGPTS P UniProtKB: Hydroxycarboxylic acid receptor 2 |
-Macromolecule #2: 8-chloranyl-3-pentyl-7H-purine-2,6-dione
| Macromolecule | Name: 8-chloranyl-3-pentyl-7H-purine-2,6-dione / type: ligand / ID: 2 / Number of copies: 1 / Formula: OKL |
|---|---|
| Molecular weight | Theoretical: 256.689 Da |
| Chemical component information |  ChemComp-OKL: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8k5d: |
 Movie
Movie Controller
Controller













 Z
Z Y
Y X
X