+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. cerevisiae Chs1 in complex with UDP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  ANTIFUNGAL PROTEIN-INHIBITOR complex ANTIFUNGAL PROTEIN-INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationchitosome / cell wall chitin biosynthetic process /  chitin synthase / chitin synthase /  chitin synthase activity / septum digestion after cytokinesis / cell septum / cell periphery / cell wall organization / chitin synthase activity / septum digestion after cytokinesis / cell septum / cell periphery / cell wall organization /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.57 Å cryo EM / Resolution: 3.57 Å | |||||||||
 Authors Authors | Bai L / Chen D | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure, catalysis, chitin transport, and selective inhibition of chitin synthase Authors: Chen DD / Wang ZB / Wang LX / Zhao P / Yun CH / Bai L | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36859.map.gz emd_36859.map.gz | 38.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36859-v30.xml emd-36859-v30.xml emd-36859.xml emd-36859.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36859.png emd_36859.png | 57.8 KB | ||
| Filedesc metadata |  emd-36859.cif.gz emd-36859.cif.gz | 6.1 KB | ||
| Others |  emd_36859_half_map_1.map.gz emd_36859_half_map_1.map.gz emd_36859_half_map_2.map.gz emd_36859_half_map_2.map.gz | 37.5 MB 37.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36859 http://ftp.pdbj.org/pub/emdb/structures/EMD-36859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36859 | HTTPS FTP |
-Related structure data
| Related structure data |  8k3tMC  8k3pC  8k3qC  8k3rC  8k3uC  8k3vC  8k3wC  8k3xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36859.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36859.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map
| File | emd_36859_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
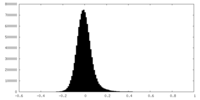
| Density Histograms |
-Half map: half map
| File | emd_36859_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : S. cerevisiae Chs1 in complex with UDP
| Entire | Name: S. cerevisiae Chs1 in complex with UDP |
|---|---|
| Components |
|
-Supramolecule #1: S. cerevisiae Chs1 in complex with UDP
| Supramolecule | Name: S. cerevisiae Chs1 in complex with UDP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
-Macromolecule #1: Chitin synthase 1
| Macromolecule | Name: Chitin synthase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number:  chitin synthase chitin synthase |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Molecular weight | Theoretical: 130.001141 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Sequence | String: MSDQNNRSRN EYHSNRKNEP SYELQNAHSG LFHSSNEELT NRNQRYTNQN ASMGSFTPVQ SLQFPEQSQQ TNMLYNGDDG NNNTINDNE RDIYGGFVNH HRQRPPPATA EYNDVFNTNS QQLPSEHQYN NVPSYPLPSI NVIQTTPELI HNGSQTMATP I ERPFFNEN ...String: MSDQNNRSRN EYHSNRKNEP SYELQNAHSG LFHSSNEELT NRNQRYTNQN ASMGSFTPVQ SLQFPEQSQQ TNMLYNGDDG NNNTINDNE RDIYGGFVNH HRQRPPPATA EYNDVFNTNS QQLPSEHQYN NVPSYPLPSI NVIQTTPELI HNGSQTMATP I ERPFFNEN DYYYNNRNSR TSPSIASSSD GYADQEARPI LEQPNNNMNS GNIPQYHDQP FGYNNGYHGL QAKDYYDDPE GG YIDQRGD DYQINSYLGR NGEMVDPYDY ENSLRHMTPM ERREYLHDDS RPVNDGKEEL DSVKSGYSHR DLGEYDKDDF SRD DEYDDL NTIDKLQFQA NGVPASSSVS SIGSKESDII VSNDNLTANR ALKRSGTEIR KFKLWNGNFV FDSPISKTLL DQYA TTTEN ANTLPNEFKF MRYQAVTCEP NQLAEKNFTV RQLKYLTPRE TELMLVVTMY NEDHILLGRT LKGIMDNVKY MVKKK NSST WGPDAWKKIV VCIISDGRSK INERSLALLS SLGCYQDGFA KDEINEKKVA MHVYEHTTMI NITNISESEV SLECNQ GTV PIQLLFCLKE QNQKKINSHR WAFEGFAELL RPNIVTLLDA GTMPGKDSIY QLWREFRNPN VGGACGEIRT DLGKRFV KL LNPLVASQNF EYKMSNILDK TTESNFGFIT VLPGAFSAYR FEAVRGQPLQ KYFYGEIMEN EGFHFFSSNM YLAEDRIL C FEVVTKKNCN WILKYCRSSY ASTDVPERVP EFILQRRRWL NGSFFASVYS FCHFYRVWSS GHNIGRKLLL TVEFFYLFF NTLISWFSLS SFFLVFRILT VSIALAYHSA FNVLSVIFLW LYGICTLSTF ILSLGNKPKS TEKFYVLTCV IFAVMMIYMI FCSIFMSVK SFQNILKNDT ISFEGLITTE AFRDIVISLG STYCLYLISS IIYLQPWHML TSFIQYILLS PSYINVLNIY A FCNVHDLS WGTKGAMANP LGKINTTEDG TFKMEVLVSS SEIQANYDKY LKVLNDFDPK SESRPTEPSY DEKKTGYYAN VR SLVIIFW VITNFIIVAV VLETGGIADY IAMKSISTDD TLETAKKAEI PLMTSKASIY FNVILWLVAL SALIRFIGCS IYM IVRFFK KVTFR UniProtKB:  Chitin synthase 1 Chitin synthase 1 |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: URIDINE-5'-DIPHOSPHATE
| Macromolecule | Name: URIDINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: UDP |
|---|---|
| Molecular weight | Theoretical: 404.161 Da |
| Chemical component information |  ChemComp-UDP: |
-Macromolecule #4: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 4 / Number of copies: 2 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.57 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 184449 |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X