+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
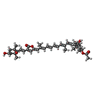
| Title | PSI-AcpPCI supercomplex from Amphidinium carterae | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | Photosysrem I /  Photosynthesis / Photosynthesis /  Dinoflagellate Dinoflagellate | |||||||||||||||||||||||||||
| Biological species |   Amphidinium carterae (eukaryote) Amphidinium carterae (eukaryote) | |||||||||||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.9 Å cryo EM / Resolution: 2.9 Å | |||||||||||||||||||||||||||
 Authors Authors | Li ZH / Li XY / Wang WD | |||||||||||||||||||||||||||
| Funding support |  China, 8 items China, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates. Authors: Xiaoyi Li / Zhenhua Li / Fangfang Wang / Songhao Zhao / Caizhe Xu / Zhiyuan Mao / Jialin Duan / Yue Feng / Yang Yang / Lili Shen / Guanglei Wang / Yanyan Yang / Long-Jiang Yu / Min Sang / ...Authors: Xiaoyi Li / Zhenhua Li / Fangfang Wang / Songhao Zhao / Caizhe Xu / Zhiyuan Mao / Jialin Duan / Yue Feng / Yang Yang / Lili Shen / Guanglei Wang / Yanyan Yang / Long-Jiang Yu / Min Sang / Guangye Han / Xuchu Wang / Tingyun Kuang / Jian-Ren Shen / Wenda Wang /   Abstract: Marine photosynthetic dinoflagellates are a group of successful phytoplankton that can form red tides in the ocean and also symbiosis with corals. These features are closely related to the ...Marine photosynthetic dinoflagellates are a group of successful phytoplankton that can form red tides in the ocean and also symbiosis with corals. These features are closely related to the photosynthetic properties of dinoflagellates. We report here three structures of photosystem I (PSI)-chlorophylls (Chls) /-peridinin protein complex (PSI-AcpPCI) from two species of dinoflagellates by single-particle cryoelectron microscopy. The crucial PsaA/B subunits of a red tidal dinoflagellate are remarkably smaller and hence losing over 20 pigment-binding sites, whereas its PsaD/F/I/J/L/M/R subunits are larger and coordinate some additional pigment sites compared to other eukaryotic photosynthetic organisms, which may compensate for the smaller PsaA/B subunits. Similar modifications are observed in a coral symbiotic dinoflagellate species, where two additional core proteins and fewer AcpPCIs are identified in the PSI-AcpPCI supercomplex. The antenna proteins AcpPCIs in dinoflagellates developed some loops and pigment sites as a result to accommodate the changed PSI core, therefore the structures of PSI-AcpPCI supercomplex of dinoflagellates reveal an unusual protein assembly pattern. A huge pigment network comprising Chls and and various carotenoids is revealed from the structural analysis, which provides the basis for our deeper understanding of the energy transfer and dissipation within the PSI-AcpPCI supercomplex, as well as the evolution of photosynthetic organisms. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36678.map.gz emd_36678.map.gz | 258.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36678-v30.xml emd-36678-v30.xml emd-36678.xml emd-36678.xml | 45.9 KB 45.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36678_fsc.xml emd_36678_fsc.xml | 16.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_36678.png emd_36678.png | 118 KB | ||
| Filedesc metadata |  emd-36678.cif.gz emd-36678.cif.gz | 10.3 KB | ||
| Others |  emd_36678_half_map_1.map.gz emd_36678_half_map_1.map.gz emd_36678_half_map_2.map.gz emd_36678_half_map_2.map.gz | 475.2 MB 475.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36678 http://ftp.pdbj.org/pub/emdb/structures/EMD-36678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36678 | HTTPS FTP |
-Related structure data
| Related structure data |  8jw0MC  8jzeC  8jzfC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36678.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36678.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36678_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36678_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : PSI-AcpPCI supercomplex from Amphidinium carterae
+Supramolecule #1: PSI-AcpPCI supercomplex from Amphidinium carterae
+Macromolecule #1: Photosystem I PsaA
+Macromolecule #2: Photosystem I PsaB
+Macromolecule #3: Photosystem I PsaC
+Macromolecule #4: Photosystem I PsaD
+Macromolecule #5: Photosystem I PsaE
+Macromolecule #6: Photosystem I PsaF
+Macromolecule #7: Photosystem I PsaR
+Macromolecule #8: Photosystem I PsaI
+Macromolecule #9: Photosystem I PsaJ
+Macromolecule #10: Photosystem I PsaL
+Macromolecule #11: Photosystem I PsaM
+Macromolecule #12: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-10, acpPCI-10
+Macromolecule #13: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-8, acpPCI-8
+Macromolecule #14: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-7, acpPCI-7
+Macromolecule #15: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-6, acpPCI-6
+Macromolecule #16: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-2, acpPCI-2
+Macromolecule #17: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-3, acpPCI-3
+Macromolecule #18: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-4, acpPCI-4
+Macromolecule #19: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-5, acpPCI-5
+Macromolecule #20: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-9, acpPCI-9
+Macromolecule #21: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-11, acpPCI-11
+Macromolecule #22: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-12, acpPCI-12
+Macromolecule #23: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-13, acpPCI-13
+Macromolecule #24: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-15, acpPCI-15
+Macromolecule #25: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-16, acpPCI-16
+Macromolecule #26: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-17, acpPCI-17
+Macromolecule #27: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-14, acpPCI-14
+Macromolecule #28: Chlorophyll a-chlorophyll c-peridinin-protein-complex I-1, acpPCI-1
+Macromolecule #29: CHLOROPHYLL A
+Macromolecule #30: PHYLLOQUINONE
+Macromolecule #31: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #32: BETA-CAROTENE
+Macromolecule #33: IRON/SULFUR CLUSTER
+Macromolecule #34: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #35: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #36: (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,be...
+Macromolecule #37: PERIDININ
+Macromolecule #38: [(1~{S},5~{R})-3,3,5-trimethyl-5-oxidanyl-4-[(3~{E},5~{E},7~{E},9...
+Macromolecule #39: Chlorophyll c1
+Macromolecule #40: 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X