+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The EREG-bound EGFR/HER2 ectodomain complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationluteinizing hormone signaling pathway / ovarian cumulus expansion /  ovulation / ERBB4-ERBB4 signaling pathway / negative regulation of smooth muscle cell differentiation / negative regulation of immature T cell proliferation in thymus / ERBB3:ERBB2 complex / ERBB2-ERBB4 signaling pathway / GRB7 events in ERBB2 signaling / immature T cell proliferation in thymus ...luteinizing hormone signaling pathway / ovarian cumulus expansion / ovulation / ERBB4-ERBB4 signaling pathway / negative regulation of smooth muscle cell differentiation / negative regulation of immature T cell proliferation in thymus / ERBB3:ERBB2 complex / ERBB2-ERBB4 signaling pathway / GRB7 events in ERBB2 signaling / immature T cell proliferation in thymus ...luteinizing hormone signaling pathway / ovarian cumulus expansion /  ovulation / ERBB4-ERBB4 signaling pathway / negative regulation of smooth muscle cell differentiation / negative regulation of immature T cell proliferation in thymus / ERBB3:ERBB2 complex / ERBB2-ERBB4 signaling pathway / GRB7 events in ERBB2 signaling / immature T cell proliferation in thymus / RNA polymerase I core binding / primary follicle stage / female meiotic nuclear division / semaphorin receptor complex / PI3K events in ERBB4 signaling / ovulation / ERBB4-ERBB4 signaling pathway / negative regulation of smooth muscle cell differentiation / negative regulation of immature T cell proliferation in thymus / ERBB3:ERBB2 complex / ERBB2-ERBB4 signaling pathway / GRB7 events in ERBB2 signaling / immature T cell proliferation in thymus / RNA polymerase I core binding / primary follicle stage / female meiotic nuclear division / semaphorin receptor complex / PI3K events in ERBB4 signaling /  oocyte maturation / regulation of microtubule-based process / ErbB-3 class receptor binding / positive regulation of innate immune response / Sema4D induced cell migration and growth-cone collapse / motor neuron axon guidance / response to hydroxyisoflavone / oocyte maturation / regulation of microtubule-based process / ErbB-3 class receptor binding / positive regulation of innate immune response / Sema4D induced cell migration and growth-cone collapse / motor neuron axon guidance / response to hydroxyisoflavone /  multivesicular body, internal vesicle lumen / neurotransmitter receptor localization to postsynaptic specialization membrane / positive regulation of prolactin secretion / negative regulation of cardiocyte differentiation / positive regulation of protein kinase C activity / diterpenoid metabolic process / Shc-EGFR complex / multivesicular body, internal vesicle lumen / neurotransmitter receptor localization to postsynaptic specialization membrane / positive regulation of prolactin secretion / negative regulation of cardiocyte differentiation / positive regulation of protein kinase C activity / diterpenoid metabolic process / Shc-EGFR complex /  mRNA transcription / mRNA transcription /  ovulation cycle / Inhibition of Signaling by Overexpressed EGFR / ovulation cycle / Inhibition of Signaling by Overexpressed EGFR /  epidermal growth factor receptor activity / EGFR interacts with phospholipase C-gamma / positive regulation of mucus secretion / response to UV-A / epidermal growth factor receptor activity / EGFR interacts with phospholipase C-gamma / positive regulation of mucus secretion / response to UV-A /  epidermal growth factor binding / PLCG1 events in ERBB2 signaling / tongue development / positive regulation of Rho protein signal transduction / midgut development / ERBB2-EGFR signaling pathway / hydrogen peroxide metabolic process / PTK6 promotes HIF1A stabilization / digestive tract morphogenesis / epidermal growth factor binding / PLCG1 events in ERBB2 signaling / tongue development / positive regulation of Rho protein signal transduction / midgut development / ERBB2-EGFR signaling pathway / hydrogen peroxide metabolic process / PTK6 promotes HIF1A stabilization / digestive tract morphogenesis /  regulation of nitric-oxide synthase activity / regulation of nitric-oxide synthase activity /  epidermal growth factor receptor binding / neuromuscular junction development / ERBB2 Activates PTK6 Signaling / morphogenesis of an epithelial fold / intracellular vesicle / Signaling by EGFR / Drug-mediated inhibition of ERBB2 signaling / Resistance of ERBB2 KD mutants to trastuzumab / Resistance of ERBB2 KD mutants to sapitinib / Resistance of ERBB2 KD mutants to tesevatinib / Resistance of ERBB2 KD mutants to neratinib / Resistance of ERBB2 KD mutants to osimertinib / Resistance of ERBB2 KD mutants to afatinib / Resistance of ERBB2 KD mutants to AEE788 / Resistance of ERBB2 KD mutants to lapatinib / epidermal growth factor receptor binding / neuromuscular junction development / ERBB2 Activates PTK6 Signaling / morphogenesis of an epithelial fold / intracellular vesicle / Signaling by EGFR / Drug-mediated inhibition of ERBB2 signaling / Resistance of ERBB2 KD mutants to trastuzumab / Resistance of ERBB2 KD mutants to sapitinib / Resistance of ERBB2 KD mutants to tesevatinib / Resistance of ERBB2 KD mutants to neratinib / Resistance of ERBB2 KD mutants to osimertinib / Resistance of ERBB2 KD mutants to afatinib / Resistance of ERBB2 KD mutants to AEE788 / Resistance of ERBB2 KD mutants to lapatinib /  Drug resistance in ERBB2 TMD/JMD mutants / Drug resistance in ERBB2 TMD/JMD mutants /  enzyme-linked receptor protein signaling pathway / positive regulation of transcription by RNA polymerase I / response to cobalamin / negative regulation of epidermal growth factor receptor signaling pathway / enzyme-linked receptor protein signaling pathway / positive regulation of transcription by RNA polymerase I / response to cobalamin / negative regulation of epidermal growth factor receptor signaling pathway /  transmembrane receptor protein tyrosine kinase activator activity / ERBB2-ERBB3 signaling pathway / protein tyrosine kinase activator activity / Signaling by ERBB4 / regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / protein insertion into membrane / eyelid development in camera-type eye / oligodendrocyte differentiation / cerebral cortex cell migration / ERBB2 Regulates Cell Motility / semaphorin-plexin signaling pathway / regulation of JNK cascade / positive regulation of cell division / activation of phospholipase C activity / positive regulation of cyclin-dependent protein serine/threonine kinase activity / PI3K events in ERBB2 signaling / positive regulation of protein targeting to membrane / keratinocyte proliferation / negative regulation of mitotic cell cycle / positive regulation of cell adhesion / hair follicle development / transmembrane receptor protein tyrosine kinase activator activity / ERBB2-ERBB3 signaling pathway / protein tyrosine kinase activator activity / Signaling by ERBB4 / regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / protein insertion into membrane / eyelid development in camera-type eye / oligodendrocyte differentiation / cerebral cortex cell migration / ERBB2 Regulates Cell Motility / semaphorin-plexin signaling pathway / regulation of JNK cascade / positive regulation of cell division / activation of phospholipase C activity / positive regulation of cyclin-dependent protein serine/threonine kinase activity / PI3K events in ERBB2 signaling / positive regulation of protein targeting to membrane / keratinocyte proliferation / negative regulation of mitotic cell cycle / positive regulation of cell adhesion / hair follicle development /  MAP kinase kinase kinase activity / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / SHC1 events in ERBB4 signaling / positive regulation of G1/S transition of mitotic cell cycle / anatomical structure morphogenesis / embryonic placenta development / positive regulation of bone resorption / positive regulation of protein kinase activity / GAB1 signalosome / MAP kinase kinase kinase activity / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / SHC1 events in ERBB4 signaling / positive regulation of G1/S transition of mitotic cell cycle / anatomical structure morphogenesis / embryonic placenta development / positive regulation of bone resorption / positive regulation of protein kinase activity / GAB1 signalosome /  regulation of angiogenesis / Nuclear signaling by ERBB4 / positive regulation of nitric oxide mediated signal transduction / regulation of angiogenesis / Nuclear signaling by ERBB4 / positive regulation of nitric oxide mediated signal transduction /  coreceptor activity coreceptor activitySimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.53 Å cryo EM / Resolution: 4.53 Å | |||||||||
 Authors Authors | Zhang Z / Bai X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2023 Journal: Cell Discov / Year: 2023Title: Structure and dynamics of the EGFR/HER2 heterodimer. Authors: Xue Bai / Pengyu Sun / Xinghao Wang / Changkun Long / Shuyun Liao / Song Dang / Shangshang Zhuang / Yongtao Du / Xinyi Zhang / Nan Li / Kangmin He / Zhe Zhang /  Abstract: HER2 belongs to the human epidermal growth factor receptor tyrosine kinase family. Its overexpression or hyperactivation is a leading cause for multiple types of cancers. HER2 functions mainly ...HER2 belongs to the human epidermal growth factor receptor tyrosine kinase family. Its overexpression or hyperactivation is a leading cause for multiple types of cancers. HER2 functions mainly through dimerization with other family members, such as EGFR. However, the molecular details for heterodimer assembly have not been completely understood. Here, we report cryo-EM structures of the EGF- and epiregulin-bound EGFR/HER2 ectodomain complexes at resolutions of 3.3 Å and 4.5 Å, respectively. Together with the functional analyses, we demonstrate that only the dimerization arm of HER2, but not that of EGFR, is essential for their heterodimer formation and signal transduction. Moreover, we analyze the differential membrane dynamics and transient interactions of endogenous EGFR and HER2 molecules in genome-edited cells using single-molecule live-cell imaging. Furthermore, we show that the interaction with HER2 could allow EGFR to resist endocytosis. Together, this work deepens our understanding of the unique structural properties and dynamics of the EGFR/HER2 complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34745.map.gz emd_34745.map.gz | 191.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34745-v30.xml emd-34745-v30.xml emd-34745.xml emd-34745.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34745.png emd_34745.png | 179 KB | ||
| Others |  emd_34745_half_map_1.map.gz emd_34745_half_map_1.map.gz emd_34745_half_map_2.map.gz emd_34745_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34745 http://ftp.pdbj.org/pub/emdb/structures/EMD-34745 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34745 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34745 | HTTPS FTP |
-Related structure data
| Related structure data |  8hgpMC  8hgoC  8hgsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34745.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34745.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34745_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
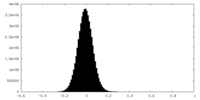
| Density Histograms |
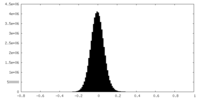
-Half map: #1
| File | emd_34745_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : EREG-bound EGFR/HER2 ectodomain complex
| Entire | Name: EREG-bound EGFR/HER2 ectodomain complex |
|---|---|
| Components |
|
-Supramolecule #1: EREG-bound EGFR/HER2 ectodomain complex
| Supramolecule | Name: EREG-bound EGFR/HER2 ectodomain complex / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Receptor tyrosine-protein kinase erbB-2
| Macromolecule | Name: Receptor tyrosine-protein kinase erbB-2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number:  receptor protein-tyrosine kinase receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 82.149758 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MELAALCRWG LLLALLPPGA ASTQVCTGTD MKLRLPASPE THLDMLRHLY QGCQVVQGNL ELTYLPTNAS LSFLQDIQEV QGYVLIAHN QVRQVPLQRL RIVRGTQLFE DNYALAVLDN GDPLNNTTPV TGASPGGLRE LQLRSLTEIL KGGVLIQRNP Q LCYQDTIL ...String: MELAALCRWG LLLALLPPGA ASTQVCTGTD MKLRLPASPE THLDMLRHLY QGCQVVQGNL ELTYLPTNAS LSFLQDIQEV QGYVLIAHN QVRQVPLQRL RIVRGTQLFE DNYALAVLDN GDPLNNTTPV TGASPGGLRE LQLRSLTEIL KGGVLIQRNP Q LCYQDTIL WKDIFHKNNQ LALTLIDTNR SRACHPCSPM CKGSRCWGES SEDCQSLTRT VCAGGCARCK GPLPTDCCHE QC AAGCTGP KHSDCLACLH FNHSGICELH CPALVTYNTD TFESMPNPEG RYTFGASCVT ACPYNYLSTD VGSCTLVCPL HNQ EVTAED GTQRCEKCSK PCARVCYGLG MEHLREVRAV TSANIQEFAG CKKIFGSLAF LPESFDGDPA SNTAPLQPEQ LQVF ETLEE ITGYLYISAW PDSLPDLSVF QNLQVIRGRI LHNGAYSLTL QGLGISWLGL RSLRELGSGL ALIHHNTHLC FVHTV PWDQ LFRNPHQALL HTANRPEDEC VGEGLACHQL CARGHCWGPG PTQCVNCSQF LRGQECVEEC RVLQGLPREY VNARHC LPC HPECQPQNGS VTCFGPEADQ CVACAHYKDP PFCVARCPSG VKPDLSYMPI WKFPDEEGAC QPCPINCTHS CVDLDDK GC PAEQRASPLT SIISAVVGIL LVVVLGVVFG ILIKRRQQKI RKYTMRRLLQ EGGSENLYFQ GGGSAQLEKE LQALEKEN A QLEWELQALE KELAQSNSLE VLFQ |
-Macromolecule #2: Epidermal growth factor receptor
| Macromolecule | Name: Epidermal growth factor receptor / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number:  receptor protein-tyrosine kinase receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 81.075297 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRPSGTAGAA LLALLAALCP ASRALEEKKV CQGTSNKLTQ LGTFEDHFLS LQRMFNNCEV VLGNLEITYV QRNYDLSFLK TIQEVAGYV LIALNTVERI PLENLQIIRG NMYYENSYAL AVLSNYDANK TGLKELPMRN LQEILHGAVR FSNNPALCNV E SIQWRDIV ...String: MRPSGTAGAA LLALLAALCP ASRALEEKKV CQGTSNKLTQ LGTFEDHFLS LQRMFNNCEV VLGNLEITYV QRNYDLSFLK TIQEVAGYV LIALNTVERI PLENLQIIRG NMYYENSYAL AVLSNYDANK TGLKELPMRN LQEILHGAVR FSNNPALCNV E SIQWRDIV SSDFLSNMSM DFQNHLGSCQ KCDPSCPNGS CWGAGEENCQ KLTKIICAQQ CSGRCRGKSP SDCCHNQCAA GC TGPRESD CLVCRKFRDE ATCKDTCPPL MLYNPTTYQM DVNPEGKYSF GATCVKKCPR NYVVTDHGSC VRACGADSYE MEE DGVRKC KKCEGPCRKV CNGIGIGEFK DSLSINATNI KHFKNCTSIS GDLHILPVAF RGDSFTHTPP LDPQELDILK TVKE ITGFL LIQAWPENRT DLHAFENLEI IRGRTKQHGQ FSLAVVSLNI TSLGLRSLKE ISDGDVIISG NKNLCYANTI NWKKL FGTS GQKTKIISNR GENSCKATGQ VCHALCSPEG CWGPEPRDCV SCRNVSRGRE CVDKCNLLEG EPREFVENSE CIQCHP ECL PQAMNITCTG RGPDNCIQCA HYIDGPHCVK TCPAGVMGEN NTLVWKYADA GHVCHLCHPN CTYGCTGPGL EGCPTNG PK IPSIATGMVG ALLLLLVVAL GIGLFMRRRH IVRKRTLRRL LGGSENLYFQ GGGSAAQLKK KLQALKKKNA QLKWKLQA L KKKLAQSNSL EVLFQ |
-Macromolecule #3: Proepiregulin
| Macromolecule | Name: Proepiregulin / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5.57841 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: VAQVSITKCS SDMNGYCLHG QCIYLVDMSQ NYCRCEVGYT GVRCEHFFL |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.8 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: NOT APPLICABLE |
|---|---|
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.53 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 273388 |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8hgp: |
 Movie
Movie Controller
Controller




















 Z
Z Y
Y X
X