+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human Pannexin1 | |||||||||
 Map data Map data | human Pannexin1 structure | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pannexin1 / ATP release / large-pore ion channel. /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationElectric Transmission Across Gap Junctions /  leak channel activity / positive regulation of interleukin-1 alpha production / bleb / wide pore channel activity / gap junction channel activity / leak channel activity / positive regulation of interleukin-1 alpha production / bleb / wide pore channel activity / gap junction channel activity /  gap junction / positive regulation of macrophage cytokine production / gap junction / positive regulation of macrophage cytokine production /  oogenesis / response to ATP ...Electric Transmission Across Gap Junctions / oogenesis / response to ATP ...Electric Transmission Across Gap Junctions /  leak channel activity / positive regulation of interleukin-1 alpha production / bleb / wide pore channel activity / gap junction channel activity / leak channel activity / positive regulation of interleukin-1 alpha production / bleb / wide pore channel activity / gap junction channel activity /  gap junction / positive regulation of macrophage cytokine production / gap junction / positive regulation of macrophage cytokine production /  oogenesis / response to ATP / monoatomic cation transport / The NLRP3 inflammasome / positive regulation of interleukin-1 beta production / response to ischemia / oogenesis / response to ATP / monoatomic cation transport / The NLRP3 inflammasome / positive regulation of interleukin-1 beta production / response to ischemia /  calcium channel activity / calcium ion transport / calcium channel activity / calcium ion transport /  actin filament binding / cell-cell signaling / actin filament binding / cell-cell signaling /  scaffold protein binding / scaffold protein binding /  protease binding / transmembrane transporter binding / protease binding / transmembrane transporter binding /  signaling receptor binding / endoplasmic reticulum membrane / structural molecule activity / signaling receptor binding / endoplasmic reticulum membrane / structural molecule activity /  endoplasmic reticulum / protein-containing complex / endoplasmic reticulum / protein-containing complex /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.75 Å cryo EM / Resolution: 3.75 Å | |||||||||
 Authors Authors | Hussain N / Penmatsa A / Vinothkumar KR | |||||||||
| Funding support |  India, 1 items India, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural insights into pore dynamics of human Pannexin isoforms Authors: Hussain N / Apotikar A / Pidathala S / Mukherjee S / Burada AP / Sikdar SK / Vinothkumar KR / Penmatsa A | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34268.map.gz emd_34268.map.gz | 167.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34268-v30.xml emd-34268-v30.xml emd-34268.xml emd-34268.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34268_fsc.xml emd_34268_fsc.xml | 13.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_34268.png emd_34268.png | 63.2 KB | ||
| Filedesc metadata |  emd-34268.cif.gz emd-34268.cif.gz | 5.7 KB | ||
| Others |  emd_34268_half_map_1.map.gz emd_34268_half_map_1.map.gz emd_34268_half_map_2.map.gz emd_34268_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34268 http://ftp.pdbj.org/pub/emdb/structures/EMD-34268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34268 | HTTPS FTP |
-Related structure data
| Related structure data |  8gtrC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34268.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34268.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human Pannexin1 structure | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map A
| File | emd_34268_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
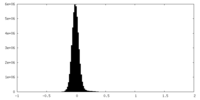
| Density Histograms |
-Half map: half map B
| File | emd_34268_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pannexin 1
| Entire | Name: Pannexin 1 PANX1 PANX1 |
|---|---|
| Components |
|
-Supramolecule #1: Pannexin 1
| Supramolecule | Name: Pannexin 1 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: human isoform 1 of Pannexin. Expressed in plasma membranes involved in ATP release |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48 kDa/nm |
-Macromolecule #1: human Pannexin1
| Macromolecule | Name: human Pannexin1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAIAQLATEY VFSDFLLKEP TEPKFKGLRL ELAVDKMVTC IAVGLPLLLI SLAFAQEISI GTQISCFSPS SFSWRQAAFV DSYCWAAVQQ KNSLQSESGN LPLWLHKFFP YILLLFAILL YLPPLFWRFA AAPHICSDLK FIMEELDKVY NRAIKAAKSA RDLDMRDGAC ...String: MAIAQLATEY VFSDFLLKEP TEPKFKGLRL ELAVDKMVTC IAVGLPLLLI SLAFAQEISI GTQISCFSPS SFSWRQAAFV DSYCWAAVQQ KNSLQSESGN LPLWLHKFFP YILLLFAILL YLPPLFWRFA AAPHICSDLK FIMEELDKVY NRAIKAAKSA RDLDMRDGAC SVPGVTENLG QSLWEVSESH FKYPIVEQYL KTKKNSNNLI IKYISCRLLT LIIILLACIY LGYYFSLSSL SDEFVCSIKS GILRNDSTVP DQFQCKLIAV GIFQLLSVIN LVVYVLLAPV VVYTLFVPFR QKTDVLKVYE ILPTFDVLHF KSEGYNDLSL YNLFLEENIS EVKSYKCLKV LENIKSSGQG IDPMLLLTNL GMIKMDVVDG KTPMSAEMRE EQGNQTAELQ GMNIDSETKA NNGEKNARQR LLDSSC UniProtKB:  Pannexin-1 Pannexin-1 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: Fresh solution containing detergent was prepared for every prep. | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK IV / Details: 3.5 s single blot. | |||||||||||||||
| Details | Sample is homoheptamer purified to homogeneity. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 3.3000000000000003 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 75000 |
| Specialist optics | Phase plate: OTHER / Spherical aberration corrector: None / Chromatic aberration corrector: None / Details: Falcon III |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 100.0 K / Max: 100.0 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 1205 / Average electron dose: 29.5 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 179.6 / Target criteria: Correlation coeficient |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)