[English] 日本語
 Yorodumi
Yorodumi- EMDB-33137: The Tet-S2 state with a pseudoknotted 4-way junction of wild-type... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 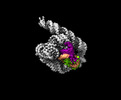 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
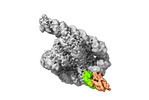
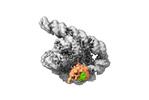
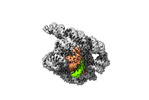
| Title | The Tet-S2 state with a pseudoknotted 4-way junction of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Biological species |   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) | ||||||||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.84 Å cryo EM / Resolution: 2.84 Å | ||||||||||||||||||||||||
 Authors Authors | Luo B / Zhang C / Ling X / Mukherjee S / Jia G / Xie J / Jia X / Liu L / Baulin EF / Luo Y ...Luo B / Zhang C / Ling X / Mukherjee S / Jia G / Xie J / Jia X / Liu L / Baulin EF / Luo Y / Jiang L / Dong H / Wei X / Bujnicki JM / Su Z | ||||||||||||||||||||||||
| Funding support |  China, China,  Poland, European Union, 7 items Poland, European Union, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Catal / Year: 2023 Journal: Nat Catal / Year: 2023Title: Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing Authors: Luo B / Zhang C / Ling X / Mukherjee S / Jia G / Xie J / Jia X / Liu L / Baulin EF / Luo Y / Jiang L / Dong H / Wei X / Bujnicki JM / Su Z | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
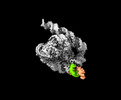
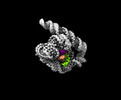
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33137.map.gz emd_33137.map.gz | 28.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33137-v30.xml emd-33137-v30.xml emd-33137.xml emd-33137.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33137.png emd_33137.png | 44.9 KB | ||
| Others |  emd_33137_half_map_1.map.gz emd_33137_half_map_1.map.gz emd_33137_half_map_2.map.gz emd_33137_half_map_2.map.gz | 33 MB 33 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33137 http://ftp.pdbj.org/pub/emdb/structures/EMD-33137 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33137 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33137 | HTTPS FTP |
-Related structure data
| Related structure data |  7xd6MC  7xd3C  7xd4C  7xd5C  7xd7C  8hd6C  8hd7C  8i7nC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33137.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33137.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33137_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33137_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Co-transcriptional folded wild-type Tetrahymena group I intron wi...
| Entire | Name: Co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon |
|---|---|
| Components |
|
-Supramolecule #1: Co-transcriptional folded wild-type Tetrahymena group I intron wi...
| Supramolecule | Name: Co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
-Macromolecule #1: The Tet-S2 state with a pseudoknotted 4-way junction molecule of ...
| Macromolecule | Name: The Tet-S2 state with a pseudoknotted 4-way junction molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (intron and 3'-exon) type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
| Molecular weight | Theoretical: 140.071703 KDa |
| Sequence | String: AUAGCAAUAU UUACCUUUGG AGGGAAAAGU UAUCAGGCAU GCACCUGGUA GCUAGUCUUU AAACCAAUAG AUUGCAUCGG UUUAAAAGG CAAGACCGUC AAAUUGCGGG AAAGGGGUCA ACAGCCGUUC AGUACCAAGU CUCAGGGGAA ACUUUGAGAU G GCCUUGCA ...String: AUAGCAAUAU UUACCUUUGG AGGGAAAAGU UAUCAGGCAU GCACCUGGUA GCUAGUCUUU AAACCAAUAG AUUGCAUCGG UUUAAAAGG CAAGACCGUC AAAUUGCGGG AAAGGGGUCA ACAGCCGUUC AGUACCAAGU CUCAGGGGAA ACUUUGAGAU G GCCUUGCA AAGGGUAUGG UAAUAAGCUG ACGGACAUGG UCCUAACCAC GCAGCCAAGU CCUAAGUCAA CAGAUCUUCU GU UGAUAUG GAUGCAGUUC ACAGACUAAA UGUCGGUCGG GGAAGAUGUA UUCUUCUCAU AAGAUAUAGU CGGACCUCUC CUU AAUGGG AGCUAGCGGA UGAAGUGAUG CAACACUGGA GCCGCUGGGA ACUAAUUUGU AUGCGAAAGU AUAUUGAUUA GUUU UGGAG UACUCGUAAG GUAGCCAAAU GCCUCGUCA |
-Macromolecule #2: The Tet-S2 state with a pseudoknotted 4-way junction molecule of ...
| Macromolecule | Name: The Tet-S2 state with a pseudoknotted 4-way junction molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon) type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
| Molecular weight | Theoretical: 8.014804 KDa |
| Sequence | String: ACGGCGGGAG UAACUAUGAC UCUCU |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 28 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: SPERMIDINE
| Macromolecule | Name: SPERMIDINE / type: ligand / ID: 4 / Number of copies: 1 / Formula: SPD |
|---|---|
| Molecular weight | Theoretical: 145.246 Da |
| Chemical component information |  ChemComp-SPD: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.00068 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.7 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 165000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.7 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 165000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 79.3 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-15 / Average electron dose: 32.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 4188150 |
|---|---|
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
| Final 3D classification | Number classes: 6 / Software - Name: RELION (ver. 3.1) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 2.84 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 494563 |
-Atomic model buiding 1
| Initial model | Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Output model |  PDB-7xd6: |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X