[English] 日本語
 Yorodumi
Yorodumi- EMDB-33112: Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 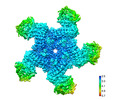 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 and the effector AvrSr35 of the wheat stem rust pathogen | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Triticum monococcum (einkorn wheat) / Triticum monococcum (einkorn wheat) /   Puccinia graminis f. sp. tritici (fungus) Puccinia graminis f. sp. tritici (fungus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.0 Å cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Alexander F / Li ET / Aaron L / Deng YN / Sun Y / Chai JJ | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: A wheat resistosome defines common principles of immune receptor channels. Authors: Alexander Förderer / Ertong Li / Aaron W Lawson / Ya-Nan Deng / Yue Sun / Elke Logemann / Xiaoxiao Zhang / Jie Wen / Zhifu Han / Junbiao Chang / Yuhang Chen / Paul Schulze-Lefert / Jijie Chai /   Abstract: Plant intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) detect pathogen effectors to trigger immune responses. Indirect recognition of a pathogen effector by the dicotyledonous ...Plant intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) detect pathogen effectors to trigger immune responses. Indirect recognition of a pathogen effector by the dicotyledonous Arabidopsis thaliana coiled-coil domain containing NLR (CNL) ZAR1 induces the formation of a large hetero-oligomeric protein complex, termed the ZAR1 resistosome, which functions as a calcium channel required for ZAR1-mediated immunity. Whether the resistosome and channel activities are conserved among plant CNLs remains unknown. Here we report the cryo-electron microscopy structure of the wheat CNL Sr35 in complex with the effector AvrSr35 of the wheat stem rust pathogen. Direct effector binding to the leucine-rich repeats of Sr35 results in the formation of a pentameric Sr35-AvrSr35 complex, which we term the Sr35 resistosome. Wheat Sr35 and Arabidopsis ZAR1 resistosomes bear striking structural similarities, including an arginine cluster in the leucine-rich repeats domain not previously recognized as conserved, which co-occurs and forms intramolecular interactions with the 'EDVID' motif in the coiled-coil domain. Electrophysiological measurements show that the Sr35 resistosome exhibits non-selective cation channel activity. These structural insights allowed us to generate new variants of closely related wheat and barley orphan NLRs that recognize AvrSr35. Our data support the evolutionary conservation of CNL resistosomes in plants and demonstrate proof of principle for structure-based engineering of NLRs for crop improvement. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33112.map.gz emd_33112.map.gz | 15.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33112-v30.xml emd-33112-v30.xml emd-33112.xml emd-33112.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33112.png emd_33112.png | 112.5 KB | ||
| Others |  emd_33112_half_map_1.map.gz emd_33112_half_map_1.map.gz emd_33112_half_map_2.map.gz emd_33112_half_map_2.map.gz | 10.1 MB 10.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33112 http://ftp.pdbj.org/pub/emdb/structures/EMD-33112 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33112 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33112 | HTTPS FTP |
-Related structure data
| Related structure data |  7xc2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33112.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33112.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33112_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
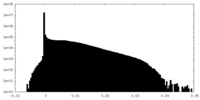
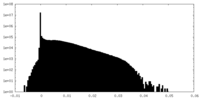
| Density Histograms |
-Half map: #2
| File | emd_33112_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 ...
| Entire | Name: Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 and the effector AvrSr35 of the wheat stem rust pathogen |
|---|---|
| Components |
|
-Supramolecule #1: Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 ...
| Supramolecule | Name: Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 and the effector AvrSr35 of the wheat stem rust pathogen type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Triticum monococcum (einkorn wheat) Triticum monococcum (einkorn wheat) |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
-Supramolecule #2: CNL9
| Supramolecule | Name: CNL9 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Puccinia graminis f. sp. tritici (fungus) Puccinia graminis f. sp. tritici (fungus) |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
-Supramolecule #3: Avirulence factor
| Supramolecule | Name: Avirulence factor / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: CNL9
| Macromolecule | Name: CNL9 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Triticum monococcum (einkorn wheat) Triticum monococcum (einkorn wheat) |
| Molecular weight | Theoretical: 102.046242 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: TITLEKKVRK GIESLITELK LMQAVLSKVS KVPADQLDEG VKIWAGNVKE LSYQMEDIVD AFMVRVGDGG ESTNPKNRVK KILKKVKKL FKNGKDLHRI SAALEEVVLQ AKQLAELRQR YEQEMRDTSA QTSVDPRMMA LYTDVTELVG IEETRDKLIN M LTEGDDWS ...String: TITLEKKVRK GIESLITELK LMQAVLSKVS KVPADQLDEG VKIWAGNVKE LSYQMEDIVD AFMVRVGDGG ESTNPKNRVK KILKKVKKL FKNGKDLHRI SAALEEVVLQ AKQLAELRQR YEQEMRDTSA QTSVDPRMMA LYTDVTELVG IEETRDKLIN M LTEGDDWS KHPLKTISIV GFGGLGKTTL AKAAYDKIKV QFDCGAFVSV SRNPEMKKVL KDILYGLDKV KYENIHNAAR DE KYLIDDI IEFLNDKRYL IVIDDIWNEK AWELIKCAFS KKSPGSRLIT TTRNVSVSEA CCSSEDDIYR MEPLSNDVSR TLF CKRIFS QEEGCPQELL KVSEEILKKC GGVPLAIITI ASLLANKGHI KAKDEWYALL SSIGHGLTKN RSLEQMKKIL LFSY YDLPS YLKPCLLYLS IFPEDREIRR ARLVWRWISE GFVYSEKQDI SLYELGDSYF NELVNRSMIQ PIGIDDEGKV KACRV HDMV LDLICSLSSE ENFVTILDDP RRKMPNSESK VRRLSIQNSK IDVDTTRMEH MRSVTVFSDN VVGKVLDISR FKVLRV LDL EGCHVSDVGY VGNLLHLRYL GLKGTHVKDL PMEVGKLQFL LTLDLRGTKI EVLPWSVVQL RRLMCLYVDY GMKLPSG IG NLTFLEVLDD LGLSDVDLDF VKELGRLTKL RVLRLDFHGF DQSMGKALEE SISNMYKLDS LDVFVNRGLI NCLSEHWV P PPRLCRLAFP SKRSWFKTLP SWINPSSLPL LSYLDITLFE VRSEDIQLLG TLPALVYLEI WNYSVFEEAH EVEAPVLSS GAALFPCATE CRFIGIGAVP SMFPQGAAPR LKRLWFTFPA KWSSSENIGL GMRHLPSLQR VVVDVISEGA SREEADEAEA ALRAAAEDH PNRPILDIW |
-Macromolecule #2: Avirulence factor
| Macromolecule | Name: Avirulence factor / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Puccinia graminis f. sp. tritici (fungus) Puccinia graminis f. sp. tritici (fungus) |
| Molecular weight | Theoretical: 50.51148 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: VNFPFPKKMI TESNSKDIRE YLASTFPFEQ QSTILDSVKS IAKVQIDDRK AFDLQLKFRQ ENLAELKDQI ILSLGANNGN QNWQKLLDY TNKLDELSNT KISPEEFIEE IQKVLYKVKL ESTSTSKLYS QFNLSIQDFA LQIIHSKYKS NQISQNDLLK L ITEDEMLK ...String: VNFPFPKKMI TESNSKDIRE YLASTFPFEQ QSTILDSVKS IAKVQIDDRK AFDLQLKFRQ ENLAELKDQI ILSLGANNGN QNWQKLLDY TNKLDELSNT KISPEEFIEE IQKVLYKVKL ESTSTSKLYS QFNLSIQDFA LQIIHSKYKS NQISQNDLLK L ITEDEMLK ILAKTKVLTY KMKYFDSASK MGINKYISTE MMDLDWQFSH YKTFNDALKK NKASDSSYLG WLTHGYSIKY GL SPNNERS MFFQDGRKYA ELYAFSKSPH RKIIPGEHLK DLLAKINKSK GIFLDQNALL DKRIYAFHEL NTLETHFPGI TSS FTDDLK SNYRKKMESV SLTCQVLQEI GNIHRFIESK VPYHSSTEYG LFSIPKIFSI PIDYKHGEKE NLVSYVDFLY STAH ERILQ DNSINQLCLD PLQESLNRIK SNIPVFFNL |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 5 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DARK FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
- Image processing
Image processing
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
|---|---|
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.33 CUT-OFF / Number images used: 230485 |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X