+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
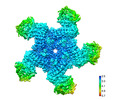
| Title | SR35-LRR with AvrSR35 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Triticum monococcum (einkorn wheat) Triticum monococcum (einkorn wheat) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.33 Å cryo EM / Resolution: 3.33 Å | |||||||||
 Authors Authors | Alexander F / Li ET / Chai JJ | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: A wheat resistosome defines common principles of immune receptor channels. Authors: Alexander Förderer / Ertong Li / Aaron W Lawson / Ya-Nan Deng / Yue Sun / Elke Logemann / Xiaoxiao Zhang / Jie Wen / Zhifu Han / Junbiao Chang / Yuhang Chen / Paul Schulze-Lefert / Jijie Chai /   Abstract: Plant intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) detect pathogen effectors to trigger immune responses. Indirect recognition of a pathogen effector by the dicotyledonous ...Plant intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) detect pathogen effectors to trigger immune responses. Indirect recognition of a pathogen effector by the dicotyledonous Arabidopsis thaliana coiled-coil domain containing NLR (CNL) ZAR1 induces the formation of a large hetero-oligomeric protein complex, termed the ZAR1 resistosome, which functions as a calcium channel required for ZAR1-mediated immunity. Whether the resistosome and channel activities are conserved among plant CNLs remains unknown. Here we report the cryo-electron microscopy structure of the wheat CNL Sr35 in complex with the effector AvrSr35 of the wheat stem rust pathogen. Direct effector binding to the leucine-rich repeats of Sr35 results in the formation of a pentameric Sr35-AvrSr35 complex, which we term the Sr35 resistosome. Wheat Sr35 and Arabidopsis ZAR1 resistosomes bear striking structural similarities, including an arginine cluster in the leucine-rich repeats domain not previously recognized as conserved, which co-occurs and forms intramolecular interactions with the 'EDVID' motif in the coiled-coil domain. Electrophysiological measurements show that the Sr35 resistosome exhibits non-selective cation channel activity. These structural insights allowed us to generate new variants of closely related wheat and barley orphan NLRs that recognize AvrSr35. Our data support the evolutionary conservation of CNL resistosomes in plants and demonstrate proof of principle for structure-based engineering of NLRs for crop improvement. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33111.map.gz emd_33111.map.gz | 6.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33111-v30.xml emd-33111-v30.xml emd-33111.xml emd-33111.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33111.png emd_33111.png | 59.7 KB | ||
| Others |  emd_33111_half_map_1.map.gz emd_33111_half_map_1.map.gz emd_33111_half_map_2.map.gz emd_33111_half_map_2.map.gz | 141.4 MB 141.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33111 http://ftp.pdbj.org/pub/emdb/structures/EMD-33111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33111 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33111.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33111.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33111_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33111_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : A wheat resistosome
| Entire | Name: A wheat resistosome |
|---|---|
| Components |
|
-Supramolecule #1: A wheat resistosome
| Supramolecule | Name: A wheat resistosome / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Triticum monococcum (einkorn wheat) Triticum monococcum (einkorn wheat) |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
-Macromolecule #1: Sr35_LRR and AvrSr35
| Macromolecule | Name: Sr35_LRR and AvrSr35 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Triticum monococcum (einkorn wheat) Triticum monococcum (einkorn wheat) |
| Sequence | String: MSHHFGLRKI KLLILLFLQV HGSQCAMRNF AADRVHGVES VISGSKSSSN PMALSKSMDK PDTSDLVDSN VQAKNDGSRY EEDFTAKYSE QVDHVSKILK EIEEQEPGTI IIDHKAFPIQ DKSPKQVVNF PFPKKMITES NSKDIREYLA STFPFEQQST ILDSVKSIAK ...String: MSHHFGLRKI KLLILLFLQV HGSQCAMRNF AADRVHGVES VISGSKSSSN PMALSKSMDK PDTSDLVDSN VQAKNDGSRY EEDFTAKYSE QVDHVSKILK EIEEQEPGTI IIDHKAFPIQ DKSPKQVVNF PFPKKMITES NSKDIREYLA STFPFEQQST ILDSVKSIAK VQIDDRKAFD LQLKFRQENL AELKDQIILS LGANNGNQNW QKLLDYTNKL DELSNTKISP EEFIEEIQKV LYKVKLESTS TSKLYSQFNL SIQDFALQII HSKYKSNQIS QNDLLKLITE DEMLKILAKT KVLTYKMKYF DSASKMGINK YISTEMMDLD WQFSHYKTFN DALKKNKASD SSYLGWLTHG YSIKYGLSPN NERSMFFQDG RKYAELYAFS KSPHRKIIPG EHLKDLLAKI NKSKGIFLDQ NALLDKRIYA FHELNTLETH FPGITSSFTD DLKSNYRKKM ESVSLTCQVL QEIGNIHRFI ESKVPYHSST EYGLFSIPKI FSIPIDYKHG EKENLVSYVD FLYSTAHERI LQDNSINQLC LDPLQESLNR IKSNIPVFFN LASHSSPIKP SNVHEGKL |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.3 µm Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.3 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.33 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 476069 |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X

















