[English] 日本語
 Yorodumi
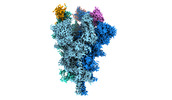
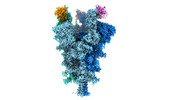
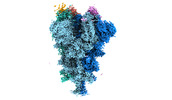
Yorodumi- EMDB-32081: Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) boun... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P17 | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | spike /  nanobody / VHH / nanobody / VHH /  VIRAL PROTEIN VIRAL PROTEIN | |||||||||||||||||||||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus 2 / Severe acute respiratory syndrome coronavirus 2 /   Vicugna pacos (alpaca) Vicugna pacos (alpaca) | |||||||||||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.29 Å cryo EM / Resolution: 3.29 Å | |||||||||||||||||||||||||||
 Authors Authors | Maeda R / Fujita J / Konishi Y / Kazuma Y / Yamazaki H / Anzai I / Yamaguchi K / Kasai K / Nagata K / Yamaoka Y ...Maeda R / Fujita J / Konishi Y / Kazuma Y / Yamazaki H / Anzai I / Yamaguchi K / Kasai K / Nagata K / Yamaoka Y / Miyakawa K / Ryo A / Shirakawa K / Makino F / Matsuura Y / Inoue T / Imura A / Namba K / Takaori-Kondo A | |||||||||||||||||||||||||||
| Funding support |  Japan, 8 items Japan, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron. Authors: Ryota Maeda / Junso Fujita / Yoshinobu Konishi / Yasuhiro Kazuma / Hiroyuki Yamazaki / Itsuki Anzai / Tokiko Watanabe / Keishi Yamaguchi / Kazuki Kasai / Kayoko Nagata / Yutaro Yamaoka / Kei ...Authors: Ryota Maeda / Junso Fujita / Yoshinobu Konishi / Yasuhiro Kazuma / Hiroyuki Yamazaki / Itsuki Anzai / Tokiko Watanabe / Keishi Yamaguchi / Kazuki Kasai / Kayoko Nagata / Yutaro Yamaoka / Kei Miyakawa / Akihide Ryo / Kotaro Shirakawa / Kei Sato / Fumiaki Makino / Yoshiharu Matsuura / Tsuyoshi Inoue / Akihiro Imura / Keiichi Namba / Akifumi Takaori-Kondo /  Abstract: We are amid the historic coronavirus infectious disease 2019 (COVID-19) pandemic. Imbalances in the accessibility of vaccines, medicines, and diagnostics among countries, regions, and populations, ...We are amid the historic coronavirus infectious disease 2019 (COVID-19) pandemic. Imbalances in the accessibility of vaccines, medicines, and diagnostics among countries, regions, and populations, and those in war crises, have been problematic. Nanobodies are small, stable, customizable, and inexpensive to produce. Herein, we present a panel of nanobodies that can detect the spike proteins of five SARS-CoV-2 variants of concern (VOCs) including Omicron. Here we show via ELISA, lateral flow, kinetic, flow cytometric, microscopy, and Western blotting assays that our nanobodies can quantify the spike variants. This panel of nanobodies broadly neutralizes viral infection caused by pseudotyped and authentic SARS-CoV-2 VOCs. Structural analyses show that the P86 clone targets epitopes that are conserved yet unclassified on the receptor-binding domain (RBD) and contacts the N-terminal domain (NTD). Human antibodies rarely access both regions; consequently, the clone buries hidden crevasses of SARS-CoV-2 spike proteins that go undetected by conventional antibodies. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32081.map.gz emd_32081.map.gz | 168 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32081-v30.xml emd-32081-v30.xml emd-32081.xml emd-32081.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32081_fsc.xml emd_32081_fsc.xml | 12.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_32081.png emd_32081.png | 56.2 KB | ||
| Masks |  emd_32081_msk_1.map emd_32081_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32081.cif.gz emd-32081.cif.gz | 6.6 KB | ||
| Others |  emd_32081_half_map_1.map.gz emd_32081_half_map_1.map.gz emd_32081_half_map_2.map.gz emd_32081_half_map_2.map.gz | 165.3 MB 165.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32081 http://ftp.pdbj.org/pub/emdb/structures/EMD-32081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32081 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32081.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32081.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.874 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_32081_msk_1.map emd_32081_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_32081_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_32081_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobod...
| Entire | Name: SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P17 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobod...
| Supramolecule | Name: SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P17 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
-Supramolecule #2: SARS-CoV-2 spike protein (2-up RBD)
| Supramolecule | Name: SARS-CoV-2 spike protein (2-up RBD) / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Vicugna pacos (alpaca) Vicugna pacos (alpaca) |
-Supramolecule #3: neutralizing nanobodies P17
| Supramolecule | Name: neutralizing nanobodies P17 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI NLVRDLPQGF SALEPLVDLP IGINITRFQT LLALHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQTGKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF PLQSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESNKKFL PFQQFGRDIA DTTDAVRDPQ TLEILDITPC SFGGVSVITP GTNTSNQVAV LYQGVNCTEV PVAIHADQLT PTWRVYSTGS NVFQTRAGCL IGAEHVNNSY ECDIPIGAGI CASYQTQTNS PGSASSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNFTI SVTTEILPVS MTKTSVDCTM YICGDSTECS NLLLQYGSFC TQLNRALTGI AVEQDKNTQE VFAQVKQIYK TPPIKDFGGF NFSQILPDPS KPSKRSFIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFGA GAALQIPFAM QMAYRFNGIG VTQNVLYENQ KLIANQFNSA IGKIQDSLSS TASALGKLQD VVNQNAQALN TLVKQLSSNF GAISSVLNDI LSRLDPPEAE VQIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKMS ECVLGQSKRV DFCGKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIVNNTVYDP LQPELDSFKE ELDKYFKNHT SPDVDLGDIS GINASVVNIQ KEIDRLNEVA KNLNESLIDL QELGKYEQGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGSH HHHHHHH GENBANK:  GENBANK: QHD43416.1 GENBANK: QHD43416.1 |
-Macromolecule #2: Neutralizing nanobody P17
| Macromolecule | Name: Neutralizing nanobody P17 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Vicugna pacos (alpaca) Vicugna pacos (alpaca) |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: QVQLQLQESG GGLVQAGGSL RLSCAASGRT SSVYNMAWFR QTPGKEREFV AAITGNGGTT LYADSVKGRL TISRGNAKNT VSLQMNVLKP DDTAVYYCAA GGWGKERNYA YWGQGTQVTV SSHHHHHH |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: GRAPHENE Details: The graphene grid was chemically oxidized and modified. | ||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV | ||||||
| Details | Mixed with 5 times molar excess of P86. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X


























