+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30539 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of Omono River virus (strain: LZ) empty capsid. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample | Capsid protein != Omono River virus Capsid protein
| |||||||||
| Function / homology | Double-stranded RNA binding motif / Double-stranded RNA binding motif / Double stranded RNA-binding domain (dsRBD) profile. / Double-stranded RNA-binding domain /  RNA binding / Main capsid protein RNA binding / Main capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Omono River virus Omono River virus | |||||||||
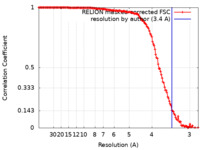
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Shao Q / Jia X / Gao Y / Liu Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2021 Journal: PLoS Pathog / Year: 2021Title: Cryo-EM reveals a previously unrecognized structural protein of a dsRNA virus implicated in its extracellular transmission. Authors: Qianqian Shao / Xudong Jia / Yuanzhu Gao / Zhe Liu / Huan Zhang / Qiqi Tan / Xin Zhang / Huiqiong Zhou / Yinyin Li / De Wu / Qinfen Zhang /  Abstract: Mosquito viruses cause unpredictable outbreaks of disease. Recently, several unassigned viruses isolated from mosquitoes, including the Omono River virus (OmRV), were identified as totivirus-like ...Mosquito viruses cause unpredictable outbreaks of disease. Recently, several unassigned viruses isolated from mosquitoes, including the Omono River virus (OmRV), were identified as totivirus-like viruses, with features similar to those of the Totiviridae family. Most reported members of this family infect fungi or protozoans and lack an extracellular life cycle stage. Here, we identified a new strain of OmRV and determined high-resolution structures for this virus using single-particle cryo-electron microscopy. The structures feature an unexpected protrusion at the five-fold vertex of the capsid. Disassociation of the protrusion could result in several conformational changes in the major capsid. All these structures, together with some biological results, suggest the protrusions' associations with the extracellular transmission of OmRV. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30539.map.gz emd_30539.map.gz | 226 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30539-v30.xml emd-30539-v30.xml emd-30539.xml emd-30539.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30539_fsc.xml emd_30539_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_30539.png emd_30539.png | 235.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30539 http://ftp.pdbj.org/pub/emdb/structures/EMD-30539 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30539 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30539 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30539.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30539.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.42 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Capsid protein
| Entire | Name: Capsid protein Capsid Capsid |
|---|---|
| Components |
|
-Supramolecule #1: Omono River virus
| Supramolecule | Name: Omono River virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 753758 / Sci species name: Omono River virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: PISADFSEVE NAPSFLSLAE NTDEVLKPYT GLEIQTIITN IVGDANPNQS RIFDQDRLRG NQYSAGGLVT QNAVSAIPFT NLIPRTIRV GNILVNSANR LQITETNVSE YYSNPIIATK LSEMISDQVK NNQFSTWRRD NTSLQGFNAF DIATINTAIL P NGLSLESM ...String: PISADFSEVE NAPSFLSLAE NTDEVLKPYT GLEIQTIITN IVGDANPNQS RIFDQDRLRG NQYSAGGLVT QNAVSAIPFT NLIPRTIRV GNILVNSANR LQITETNVSE YYSNPIIATK LSEMISDQVK NNQFSTWRRD NTSLQGFNAF DIATINTAIL P NGLSLESM LLKLSLLHSI KAMNVDAASI NRSQYQVIDH NTVPTIGAPA VVGVNNSPVF GEDCGGNNPV YPFGGGTGAI AF HVTLQTV PDERKSYAIF VPPAILQATS DANEALALFA LSMSEWPHAL YTVTKQTTDL AGANAGQQVF IPTQSTIHIG GRR VLDLII PRREIAPNPT TLVAANAMCM VRPQAGPDAT AGAIPLAAGQ LFNMNFIGAP AFEEWPMTSY LYSWAGRFDI TTIR QYMGR LATMVGVKDA YWAAHELNVA LSQVAPKMTT AAGGWAAQAA NSAQQSDVCY SSLLTVTRSA ANFPLANQPA ADMRV YDTD PATWNKVALG LATAANLVPE QSMDVPFVVG DARASFWERL QAIPMCIAWT MYYHSRGITT LAWDNAYTDN TNKWLQ KMV RNTFSTTQSV GTIIPARYGK IVCNLYKNMF HRAPAYVATS VGGKELHITH FERWLPGGTY ANVYSGAGAV VNCFSPV LI PDIWCQYFTA KLPLFAGAFP PAQGQNSTKG FNSKQGLMIH RNQNNNLVAP YLEKFADNSS YFPVGQGPEI NDMATWNG R LWMTTGNVQY LDYSGAAIVE AVPPAGELPV GKQIPLLAGE NAPIELTNAA TTCVPRYSND GRRIFTYLTT AQSVIPVQA CNRAANLARS CWLLSNVYAE PALQALGDEV EDAFDTLTNS SFLDVAKSVA ESAGEVPATK ALTDLQAVDV SSLPSTSDPS NVLSQPAPL MSPPTSSS |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller