+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of hAQP2 in DDM | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | channel /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to water deprivation / renal water transport / glycerol transmembrane transporter activity / Passive transport by Aquaporins / glycerol transmembrane transport / lumenal side of membrane / water transmembrane transporter activity / cellular response to mercury ion /  water channel activity / water transport ...cellular response to water deprivation / renal water transport / glycerol transmembrane transporter activity / Passive transport by Aquaporins / glycerol transmembrane transport / lumenal side of membrane / water transmembrane transporter activity / cellular response to mercury ion / water channel activity / water transport ...cellular response to water deprivation / renal water transport / glycerol transmembrane transporter activity / Passive transport by Aquaporins / glycerol transmembrane transport / lumenal side of membrane / water transmembrane transporter activity / cellular response to mercury ion /  water channel activity / water transport / metanephric collecting duct development / renal water homeostasis / transport vesicle membrane / cellular response to copper ion / actin filament organization / recycling endosome / Vasopressin regulates renal water homeostasis via Aquaporins / protein homotetramerization / basolateral plasma membrane / apical plasma membrane / perinuclear region of cytoplasm / water channel activity / water transport / metanephric collecting duct development / renal water homeostasis / transport vesicle membrane / cellular response to copper ion / actin filament organization / recycling endosome / Vasopressin regulates renal water homeostasis via Aquaporins / protein homotetramerization / basolateral plasma membrane / apical plasma membrane / perinuclear region of cytoplasm /  Golgi apparatus / extracellular exosome / Golgi apparatus / extracellular exosome /  membrane / membrane /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
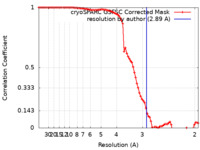
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.89 Å cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Kamegawa A / Suzuki S / Nishikawa K / Numoto N / Suzuki H / Fujiyoshi Y | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2023 Journal: J Struct Biol / Year: 2023Title: Structural analysis of the water channel AQP2 by single-particle cryo-EM. Authors: Akiko Kamegawa / Shota Suzuki / Hiroshi Suzuki / Kouki Nishikawa / Nobutaka Numoto / Yoshinori Fujiyoshi /  Abstract: Water channels, which are small membrane proteins almost entirely buried in lipid membranes, are challenging research targets for single-particle cryo-electron microscopy (cryo-EM), a powerful ...Water channels, which are small membrane proteins almost entirely buried in lipid membranes, are challenging research targets for single-particle cryo-electron microscopy (cryo-EM), a powerful technique routinely used to determine the structures of membrane proteins. Because the single-particle method enables structural analysis of a whole protein with flexible parts that interfere with crystallization, we have focused our efforts on analyzing water channel structures. Here, utilizing this system, we analyzed the structure of full-length aquaporin-2 (AQP2), a primary regulator of vasopressin-dependent reabsorption of water at the renal collecting ducts. The 2.9 Å resolution map revealed a cytoplasmic extension of the cryo-EM density that was presumed to be the highly flexible C-terminus at which the localization of AQP2 is regulated in the renal collecting duct cells. We also observed a continuous density along the common water pathway inside the channel pore and lipid-like molecules at the membrane interface. Observations of these constructions in the AQP2 structure analyzed without any fiducial markers (e.g., a rigidly bound antibody) indicate that single-particle cryo-EM will be useful for investigating water channels in native states as well as in complexes with chemical compounds. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29934.map.gz emd_29934.map.gz | 3.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29934-v30.xml emd-29934-v30.xml emd-29934.xml emd-29934.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29934_fsc.xml emd_29934_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_29934.png emd_29934.png | 57.8 KB | ||
| Masks |  emd_29934_msk_1.map emd_29934_msk_1.map | 4.3 MB |  Mask map Mask map | |
| Others |  emd_29934_half_map_1.map.gz emd_29934_half_map_1.map.gz emd_29934_half_map_2.map.gz emd_29934_half_map_2.map.gz | 3.8 MB 3.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29934 http://ftp.pdbj.org/pub/emdb/structures/EMD-29934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29934 | HTTPS FTP |
-Related structure data
| Related structure data |  8gclMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29934.map.gz / Format: CCP4 / Size: 4.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29934.map.gz / Format: CCP4 / Size: 4.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.96 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29934_msk_1.map emd_29934_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29934_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29934_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : tetrameter of hAQP2
| Entire | Name: tetrameter of hAQP2 |
|---|---|
| Components |
|
-Supramolecule #1: tetrameter of hAQP2
| Supramolecule | Name: tetrameter of hAQP2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Aquaporin-2
| Macromolecule | Name: Aquaporin-2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.862389 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MWELRSIAFS RAVFAEFLAT LLFVFFGLGS ALNWPQALPS VLQIAMAFGL GIGTLVQALG HISGAHINPA VTVACLVGCH VSVLRAAFY VAAQLLGAVA GAALLHEITP ADIRGDLAVN ALSNSTTAGQ AVTVELFLTL QLVLCIFAST DERRGENPGT P ALSIGFSV ...String: MWELRSIAFS RAVFAEFLAT LLFVFFGLGS ALNWPQALPS VLQIAMAFGL GIGTLVQALG HISGAHINPA VTVACLVGCH VSVLRAAFY VAAQLLGAVA GAALLHEITP ADIRGDLAVN ALSNSTTAGQ AVTVELFLTL QLVLCIFAST DERRGENPGT P ALSIGFSV ALGHLLGIHY TGCSMNPARS LAPAVVTGKF DDHWVFWIGP LVGAILGSLL YNYVLFPPAK SLSERLAVLK GL EPDTDWE EREVRRRQSV ELHSPQSLPR GTKA |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7.2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: MOLYBDENUM / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 69.6 e/Å2 |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X