[English] 日本語
 Yorodumi
Yorodumi- EMDB-29383: Structure of baseplate with receptor binding complex of Agrobacte... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 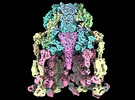 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of baseplate with receptor binding complex of Agrobacterium phage Milano | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Myophage / redox trigger /  disulfides / disulfides /  VIRUS VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Agrobacterium phage Milano (virus) Agrobacterium phage Milano (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Sonani RR / Leiman PG / Wang F / Kreutzberger MAB / Sebastian A / Esteves NC / Kelly RJ / Scharf B / Egelman EH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano. Authors: Ravi R Sonani / Lee K Palmer / Nathaniel C Esteves / Abigail A Horton / Amanda L Sebastian / Rebecca J Kelly / Fengbin Wang / Mark A B Kreutzberger / William K Russell / Petr G Leiman / ...Authors: Ravi R Sonani / Lee K Palmer / Nathaniel C Esteves / Abigail A Horton / Amanda L Sebastian / Rebecca J Kelly / Fengbin Wang / Mark A B Kreutzberger / William K Russell / Petr G Leiman / Birgit E Scharf / Edward H Egelman /  Abstract: A contractile sheath and rigid tube assembly is a widespread apparatus used by bacteriophages, tailocins, and the bacterial type VI secretion system to penetrate cell membranes. In this mechanism, ...A contractile sheath and rigid tube assembly is a widespread apparatus used by bacteriophages, tailocins, and the bacterial type VI secretion system to penetrate cell membranes. In this mechanism, contraction of an external sheath powers the motion of an inner tube through the membrane. The structure, energetics, and mechanism of the machinery imply rigidity and straightness. The contractile tail of Agrobacterium tumefaciens bacteriophage Milano is flexible and bent to varying degrees, which sets it apart from other contractile tail-like systems. Here, we report structures of the Milano tail including the sheath-tube complex, baseplate, and putative receptor-binding proteins. The flexible-to-rigid transformation of the Milano tail upon contraction can be explained by unique electrostatic properties of the tail tube and sheath. All components of the Milano tail, including sheath subunits, are crosslinked by disulfides, some of which must be reduced for contraction to occur. The putative receptor-binding complex of Milano contains a tailspike, a tail fiber, and at least two small proteins that form a garland around the distal ends of the tailspikes and tail fibers. Despite being flagellotropic, Milano lacks thread-like tail filaments that can wrap around the flagellum, and is thus likely to employ a different binding mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29383.map.gz emd_29383.map.gz | 65.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29383-v30.xml emd-29383-v30.xml emd-29383.xml emd-29383.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29383_fsc.xml emd_29383_fsc.xml | 19.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_29383.png emd_29383.png | 127.2 KB | ||
| Filedesc metadata |  emd-29383.cif.gz emd-29383.cif.gz | 7.9 KB | ||
| Others |  emd_29383_half_map_1.map.gz emd_29383_half_map_1.map.gz emd_29383_half_map_2.map.gz emd_29383_half_map_2.map.gz | 682.8 MB 682.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29383 http://ftp.pdbj.org/pub/emdb/structures/EMD-29383 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29383 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29383 | HTTPS FTP |
-Related structure data
| Related structure data |  8fqcMC  8fopC  8fouC  8foyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29383.map.gz / Format: CCP4 / Size: 736.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29383.map.gz / Format: CCP4 / Size: 736.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29383_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29383_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Agrobacterium phage Milano
+Supramolecule #1: Agrobacterium phage Milano
+Macromolecule #1: Baseplate hub protein, gp26
+Macromolecule #2: Tail-tube, gp21
+Macromolecule #3: Baseplate Wedge 2 protein, gp29
+Macromolecule #4: Baseplate wedge 1, gp28
+Macromolecule #5: Tail sheath protein, gp20
+Macromolecule #6: Baseplate Wedge 3 protein, gp30
+Macromolecule #7: Tail Spike protein, gp124
+Macromolecule #8: Short Tail Fibers, gp31
+Macromolecule #9: Baseplate Centerpiece, gp25
+Macromolecule #10: Baseplate Central Spike, gp27
+Macromolecule #11: FE (III) ION
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



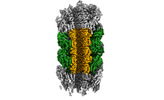


 Z
Z Y
Y X
X

















 Agrobacterium fabrum str. C58 (bacteria)
Agrobacterium fabrum str. C58 (bacteria)
