[English] 日本語
 Yorodumi
Yorodumi- EMDB-29073: Human IFT-A complex structures provide molecular insights into ci... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 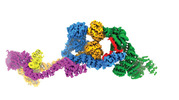 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
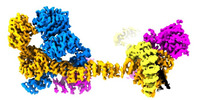
| Title | Human IFT-A complex structures provide molecular insights into ciliary transport | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsmoothened signaling pathway involved in dorsal/ventral neural tube patterning / myotome development / protein localization to non-motile cilium / regulation of intraciliary retrograde transport / : / forebrain dorsal/ventral pattern formation / ear morphogenesis / intraciliary transport particle A / intraciliary anterograde transport / cone photoreceptor outer segment ...smoothened signaling pathway involved in dorsal/ventral neural tube patterning / myotome development / protein localization to non-motile cilium / regulation of intraciliary retrograde transport / : / forebrain dorsal/ventral pattern formation / ear morphogenesis / intraciliary transport particle A / intraciliary anterograde transport / cone photoreceptor outer segment / negative regulation of eating behavior /  digestive system development / embryonic heart tube left/right pattern formation / embryonic body morphogenesis / photoreceptor cell outer segment organization / neural tube patterning / intraciliary retrograde transport / protein localization to ciliary membrane / cerebellar Purkinje cell differentiation / embryonic camera-type eye development / intraciliary transport / establishment of protein localization to organelle / gonad development / digestive system development / embryonic heart tube left/right pattern formation / embryonic body morphogenesis / photoreceptor cell outer segment organization / neural tube patterning / intraciliary retrograde transport / protein localization to ciliary membrane / cerebellar Purkinje cell differentiation / embryonic camera-type eye development / intraciliary transport / establishment of protein localization to organelle / gonad development /  regulation of cilium assembly / spinal cord dorsal/ventral patterning / photoreceptor connecting cilium / ciliary tip / regulation of cilium assembly / spinal cord dorsal/ventral patterning / photoreceptor connecting cilium / ciliary tip /  ventricular system development / ventricular system development /  Intraflagellar transport / camera-type eye morphogenesis / embryonic brain development / protein localization to cilium / non-motile cilium assembly / non-motile cilium / embryonic heart tube development / regulation of smoothened signaling pathway / embryonic cranial skeleton morphogenesis / Intraflagellar transport / camera-type eye morphogenesis / embryonic brain development / protein localization to cilium / non-motile cilium assembly / non-motile cilium / embryonic heart tube development / regulation of smoothened signaling pathway / embryonic cranial skeleton morphogenesis /  nervous system process / nervous system process /  motile cilium / embryonic forelimb morphogenesis / determination of left/right symmetry / embryonic limb morphogenesis / motile cilium / embryonic forelimb morphogenesis / determination of left/right symmetry / embryonic limb morphogenesis /  limb development / smoothened signaling pathway / embryonic digit morphogenesis / receptor clustering / limb development / smoothened signaling pathway / embryonic digit morphogenesis / receptor clustering /  axoneme / Bergmann glial cell differentiation / axoneme / Bergmann glial cell differentiation /  cilium assembly / centriolar satellite / photoreceptor outer segment / Hedgehog 'off' state / negative regulation of smoothened signaling pathway / cilium assembly / centriolar satellite / photoreceptor outer segment / Hedgehog 'off' state / negative regulation of smoothened signaling pathway /  centriole / cellular response to leukemia inhibitory factor / ciliary basal body / neural tube closure / cell morphogenesis / centriole / cellular response to leukemia inhibitory factor / ciliary basal body / neural tube closure / cell morphogenesis /  cilium / positive regulation of canonical Wnt signaling pathway / microtubule cytoskeleton / cilium / positive regulation of canonical Wnt signaling pathway / microtubule cytoskeleton /  heart development / protein-containing complex assembly / in utero embryonic development / heart development / protein-containing complex assembly / in utero embryonic development /  cytoskeleton / cytoskeleton /  centrosome / centrosome /  chromatin binding / positive regulation of gene expression / regulation of transcription by RNA polymerase II / chromatin binding / positive regulation of gene expression / regulation of transcription by RNA polymerase II /  membrane / membrane /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Jiang M / Palicharla VR / Miller D / Hwang SH / Zhu H / Hixson P / Mukhopadhyay S / Sun J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Human IFT-A complex structures provide molecular insights into ciliary transport. Authors: Meiqin Jiang / Vivek Reddy Palicharla / Darcie Miller / Sun-Hee Hwang / Hanwen Zhu / Patricia Hixson / Saikat Mukhopadhyay / Ji Sun /  Abstract: Intraflagellar transport (IFT) complexes, IFT-A and IFT-B, form bidirectional trains that move along the axonemal microtubules and are essential for assembling and maintaining cilia. Mutations in IFT ...Intraflagellar transport (IFT) complexes, IFT-A and IFT-B, form bidirectional trains that move along the axonemal microtubules and are essential for assembling and maintaining cilia. Mutations in IFT subunits lead to numerous ciliopathies involving multiple tissues. However, how IFT complexes assemble and mediate cargo transport lacks mechanistic understanding due to missing high-resolution structural information of the holo-complexes. Here we report cryo-EM structures of human IFT-A complexes in the presence and absence of TULP3 at overall resolutions of 3.0-3.9 Å. IFT-A adopts a "lariat" shape with interconnected core and peripheral subunits linked by structurally vital zinc-binding domains. TULP3, the cargo adapter, interacts with IFT-A through its N-terminal region, and interface mutations disrupt cargo transport. We also determine the molecular impacts of disease mutations on complex formation and ciliary transport. Our work reveals IFT-A architecture, sheds light on ciliary transport and IFT train formation, and enables the rationalization of disease mutations in ciliopathies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29073.map.gz emd_29073.map.gz | 350.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29073-v30.xml emd-29073-v30.xml emd-29073.xml emd-29073.xml | 32.6 KB 32.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29073.png emd_29073.png | 75.9 KB | ||
| Others |  emd_29073_additional_1.map.gz emd_29073_additional_1.map.gz emd_29073_additional_2.map.gz emd_29073_additional_2.map.gz emd_29073_additional_3.map.gz emd_29073_additional_3.map.gz emd_29073_additional_4.map.gz emd_29073_additional_4.map.gz emd_29073_half_map_1.map.gz emd_29073_half_map_1.map.gz emd_29073_half_map_2.map.gz emd_29073_half_map_2.map.gz | 389.3 MB 390.3 MB 202.9 MB 203.1 MB 348 MB 375.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29073 http://ftp.pdbj.org/pub/emdb/structures/EMD-29073 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29073 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29073 | HTTPS FTP |
-Related structure data
| Related structure data |  8fgwMC  8fh3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29073.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29073.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.48 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #4
| File | emd_29073_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #3
| File | emd_29073_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_29073_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_29073_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29073_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29073_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Intraflagellar transport complex A (IFT-A)
| Entire | Name: Intraflagellar transport complex A (IFT-A) |
|---|---|
| Components |
|
-Supramolecule #1: Intraflagellar transport complex A (IFT-A)
| Supramolecule | Name: Intraflagellar transport complex A (IFT-A) / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: WD repeat-containing protein 35
| Macromolecule | Name: WD repeat-containing protein 35 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 133.705641 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFFYLSKKIS IPNNVKLQCV SWNKEQGFIA CGGEDGLLKV LKLETQTDDA KLRGLAAPSN LSMNQTLEGH SGSVQVVTWN EQYQKLTTS DENGLIIVWM LYKGSWIEEM INNRNKSVVR SMSWNADGQK ICIVYEDGAV IVGSVDGNRI WGKDLKGIQL S HVTWSADS ...String: MFFYLSKKIS IPNNVKLQCV SWNKEQGFIA CGGEDGLLKV LKLETQTDDA KLRGLAAPSN LSMNQTLEGH SGSVQVVTWN EQYQKLTTS DENGLIIVWM LYKGSWIEEM INNRNKSVVR SMSWNADGQK ICIVYEDGAV IVGSVDGNRI WGKDLKGIQL S HVTWSADS KVLLFGMANG EIHIYDNQGN FMIKMKLSCL VNVTGAISIA GIHWYHGTEG YVEPDCPCLA VCFDNGRCQI MR HENDQNP VLIDTGMYVV GIQWNHMGSV LAVAGFQKAA MQDKDVNIVQ FYTPFGEHLG TLKVPGKEIS ALSWEGGGLK IAL AVDSFI YFANIRPNYK WGYCSNTVVY AYTRPDRPEY CVVFWDTKNN EKYVKYVKGL ISITTCGDFC ILATKADENH PQEE NEMET FGATFVLVLC NSIGTPLDPK YIDIVPLFVA MTKTHVIAAS KEAFYTWQYR VAKKLTALEI NQITRSRKEG RERIY HVDD TPSGSMDGVL DYSKTIQGTR DPICAITASD KILIVGRESG TIQRYSLPNV GLIQKYSLNC RAYQLSLNCN SSRLAI IDI SGVLTFFDLD ARVTDSTGQQ VVGELLKLER RDVWDMKWAK DNPDLFAMME KTRMYVFRNL DPEEPIQTSG YICNFED LE IKSVLLDEIL KDPEHPNKDY LINFEIRSLR DSRALIEKVG IKDASQFIED NPHPRLWRLL AEAALQKLDL YTAEQAFV R CKDYQGIKFV KRLGKLLSES MKQAEVVGYF GRFEEAERTY LEMDRRDLAI GLRLKLGDWF RVLQLLKTGS GDADDSLLE QANNAIGDYF ADRQKWLNAV QYYVQGRNQE RLAECYYMLE DYEGLENLAI SLPENHKLLP EIAQMFVRVG MCEQAVTAFL KCSQPKAAV DTCVHLNQWN KAVELAKNHS MKEIGSLLAR YASHLLEKNK TLDAIELYRK ANYFFDAAKL MFKIADEEAK K GSKPLRVK KLYVLSALLI EQYHEQMKNA QRGKVKGKSS EATSALAGLL EEEVLSTTDR FTDNAWRGAE AYHFFILAQR QL YEGCVDT ALKTALHLKD YEDIIPPVEI YSLLALCACA SRAFGTCSKA FIKLKSLETL SSEQKQQYED LALEIFTKHT SKD NRKPEL DSLMEGGEGK LPTCVATGSP ITEYQFWMCS VCKHGVLAQE ISHYSFCPLC HSPVG |
-Macromolecule #2: Intraflagellar transport protein 122 homolog
| Macromolecule | Name: Intraflagellar transport protein 122 homolog / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 141.993703 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRAVLTWRDK AEHCINDIAF KPDGTQLILA AGSRLLVYDT SDGTLLQPLK GHKDTVYCVA YAKDGKRFAS GSADKSVIIW TSKLEGILK YTHNDAIQCV SYNPITHQLA SCSSSDFGLW SPEQKSVSKH KSSSKIICCS WTNDGQYLAL GMFNGIISIR N KNGEEKVK ...String: MRAVLTWRDK AEHCINDIAF KPDGTQLILA AGSRLLVYDT SDGTLLQPLK GHKDTVYCVA YAKDGKRFAS GSADKSVIIW TSKLEGILK YTHNDAIQCV SYNPITHQLA SCSSSDFGLW SPEQKSVSKH KSSSKIICCS WTNDGQYLAL GMFNGIISIR N KNGEEKVK IERPGGSLSP IWSICWNPSS RWESFWMNRE NEDAEDVIVN RYIQEIPSTL KSAVYSSQGS EAEEEEPEED DD SPRDDNL EERNDILAVA DWGQKVSFYQ LSGKQIGKDR ALNFDPCCIS YFTKGEYILL GGSDKQVSLF TKDGVRLGTV GEQ NSWVWT CQAKPDSNYV VVGCQDGTIS FYQLIFSTVH GLYKDRYAYR DSMTDVIVQH LITEQKVRIK CKELVKKIAI YRNR LAIQL PEKILIYELY SEDLSDMHYR VKEKIIKKFE CNLLVVCANH IILCQEKRLQ CLSFSGVKER EWQMESLIRY IKVIG GPPG REGLLVGLKN GQILKIFVDN LFAIVLLKQA TAVRCLDMSA SRKKLAVVDE NDTCLVYDID TKELLFQEPN ANSVAW NTQ CEDMLCFSGG GYLNIKASTF PVHRQKLQGF VVGYNGSKIF CLHVFSISAV EVPQSAPMYQ YLDRKLFKEA YQIACLG VT DTDWRELAME ALEGLDFETA KKAFIRVQDL RYLELISSIE ERKKRGETNN DLFLADVFSY QGKFHEAAKL YKRSGHEN L ALEMYTDLCM FEYAKDFLGS GDPKETKMLI TKQADWARNI KEPKAAVEMY ISAGEHVKAI EICGDHGWVD MLIDIARKL DKAEREPLLL CATYLKKLDS PGYAAETYLK MGDLKSLVQL HVETQRWDEA FALGEKHPEF KDDIYMPYAQ WLAENDRFEE AQKAFHKAG RQREAVQVLE QLTNNAVAES RFNDAAYYYW MLSMQCLDIA QDPAQKDTML GKFYHFQRLA ELYHGYHAIH R HTEDPFSV HRPETLFNIS RFLLHSLPKD TPSGISKVKI LFTLAKQSKA LGAYRLARHA YDKLRGLYIP ARFQKSIELG TL TIRAKPF HDSEELVPLC YRCSTNNPLL NNLGNVCINC RQPFIFSASS YDVLHLVEFY LEEGITDEEA ISLIDLEVLR PKR DDRQLE IANNSSQILR LVETKDSIGD EDPFTAKLSF EQGGSEFVPV VVSRLVLRSM SRRDVLIKRW PPPLRWQYFR SLLP DASIT MCPSCFQMFH SEDYELLVLQ HGCCPYCRRC KDDPGP |
-Macromolecule #3: WD repeat-containing protein 19
| Macromolecule | Name: WD repeat-containing protein 19 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 151.760391 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKRIFSLLEK TWLGAPIQFA WQKTSGNYLA VTGADYIVKI FDRHGQKRSE INLPGNCVAM DWDKDGDVLA VIAEKSSCIY LWDANTNKT SQLDNGMRDQ MSFLLWSKVG SFLAVGTVKG NLLIYNHQTS RKIPVLGKHT KRITCGCWNA ENLLALGGED K MITVSNQE ...String: MKRIFSLLEK TWLGAPIQFA WQKTSGNYLA VTGADYIVKI FDRHGQKRSE INLPGNCVAM DWDKDGDVLA VIAEKSSCIY LWDANTNKT SQLDNGMRDQ MSFLLWSKVG SFLAVGTVKG NLLIYNHQTS RKIPVLGKHT KRITCGCWNA ENLLALGGED K MITVSNQE GDTIRQTQVR SEPSNMQFFL MKMDDRTSAA ESMISVVLGK KTLFFLNLNE PDNPADLEFQ QDFGNIVCYN WY GDGRIMI GFSCGHFVVI STHTGELGQE IFQARNHKDN LTSIAVSQTL NKVATCGDNC IKIQDLVDLK DMYVILNLDE ENK GLGTLS WTDDGQLLAL STQRGSLHVF LTKLPILGDA CSTRIAYLTS LLEVTVANPV EGELPITVSV DVEPNFVAVG LYHL AVGMN NRAWFYVLGE NAVKKLKDME YLGTVASICL HSDYAAALFE GKVQLHLIES EILDAQEERE TRLFPAVDDK CRILC HALT SDFLIYGTDT GVVQYFYIED WQFVNDYRHP VSVKKIFPDP NGTRLVFIDE KSDGFVYCPV NDATYEIPDF SPTIKG VLW ENWPMDKGVF IAYDDDKVYT YVFHKDTIQG AKVILAGSTK VPFAHKPLLL YNGELTCQTQ SGKVNNIYLS THGFLSN LK DTGPDELRPM LAQNLMLKRF SDAWEMCRIL NDEAAWNELA RACLHHMEVE FAIRVYRRIG NVGIVMSLEQ IKGIEDYN L LAGHLAMFTN DYNLAQDLYL ASSCPIAALE MRRDLQHWDS ALQLAKHLAP DQIPFISKEY AIQLEFAGDY VNALAHYEK GITGDNKEHD EACLAGVAQM SIRMGDIRRG VNQALKHPSR VLKRDCGAIL ENMKQFSEAA QLYEKGLYYD KAASVYIRSK NWAKVGDLL PHVSSPKIHL QYAKAKEADG RYKEAVVAYE NAKQWQSVIR IYLDHLNNPE KAVNIVRETQ SLDGAKMVAR F FLQLGDYG SAIQFLVMSK CNNEAFTLAQ QHNKMEIYAD IIGSEDTTNE DYQSIALYFE GEKRYLQAGK FFLLCGQYSR AL KHFLKCP SSEDNVAIEM AIETVGQAKD ELLTNQLIDH LLGENDGMPK DAKYLFRLYM ALKQYREAAQ TAIIIAREEQ SAG NYRNAH DVLFSMYAEL KSQKIKIPSE MATNLMILHS YILVKIHVKN GDHMKGARML IRVANNISKF PSHIVPILTS TVIE CHRAG LKNSAFSFAA MLMRPEYRSK IDAKYKKKIE GMVRRPDISE IEEATTPCPF CKFLLPECEL LCPGCKNSIP YCIAT GRHM LKDDWTVCPH CDFPALYSEL KIMLNTESTC PMCSERLNAA QLKKISDCTQ YLRTEEEL |
-Macromolecule #4: Tetratricopeptide repeat protein 21B
| Macromolecule | Name: Tetratricopeptide repeat protein 21B / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 151.137438 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDSQELKTLI NYYCQERYFH HVLLVASEGI KRYGSDPVFR FYHAYGTLME GKTQEALREF EAIKNKQDVS LCSLLALIYA HKMSPNPDR EAILESDARV KEQRKGAGEK ALYHAGLFLW HIGRHDKARE YIDRMIKISD GSKQGHVLKA WLDITRGKEP Y TKKALKYF ...String: MDSQELKTLI NYYCQERYFH HVLLVASEGI KRYGSDPVFR FYHAYGTLME GKTQEALREF EAIKNKQDVS LCSLLALIYA HKMSPNPDR EAILESDARV KEQRKGAGEK ALYHAGLFLW HIGRHDKARE YIDRMIKISD GSKQGHVLKA WLDITRGKEP Y TKKALKYF EEGLQDGNDT FALLGKAQCL EMRQNYSGAL ETMNQIIVNF PSFLPAFVKK MKLQLALQDW DQTVETAQRL LL QDSQNVE ALRMQALYYV CREGDIEKAS TKLENLGNAL DAMEPQNAQL FYNITLAFSR TCGRSQLILQ KIQTLLERAF SLN PQQSEF ATELGYQMIL QGRVKEALKW YKTAMTLDET SVSALVGFIQ CQLIEGQLQD ADQQLEFLNE IQQSIGKSAE LIYL HAVLA MKKNKRQEEV INLLNDVLDT HFSQLEGLPL GIQYFEKLNP DFLLEIVMEY LSFCPMQPAS PGQPLCPLLR RCISV LETV VRTVPGLLQT VFLIAKVKYL SGDIEAAFNN LQHCLEHNPS YADAHLLLAQ VYLSQEKVKL CSQSLELCLS YDFKVR DYP LYHLIKAQSQ KKMGEIADAI KTLHMAMSLP GMKRIGASTK SKDRKTEVDT SHRLSIFLEL IDVHRLNGEQ HEATKVL QD AIHEFSGTSE EVRVTIANAD LALAQGDIER ALSILQNVTA EQPYFIEARE KMADIYLKHR KDKMLYITCF REIAERMA N PRSFLLLGDA YMNILEPEEA IVAYEQALNQ NPKDGTLASK MGKALIKTHN YSMAITYYEA ALKTGQKNYL CYDLAELLL KLKWYDKAEK VLQHALAHEP VNELSALMED GRCQVLLAKV YSKMEKLGDA ITALQQAREL QARVLKRVQM EQPDAVPAQK HLAAEICAE IAKHSVAQRD YEKAIKFYRE ALVHCETDNK IMLELARLYL AQDDPDSCLR QCALLLQSDQ DNEAATMMMA D LMFRKQDY EQAVFHLQQL LERKPDNYMT LSRLIDLLRR CGKLEDVPRF FSMAEKRNSR AKLEPGFQYC KGLYLWYTGE PN DALRHFN KARKDRDWGQ NALYNMIEIC LNPDNETVGG EVFENLDGDL GNSTEKQESV QLAVRTAEKL LKELKPQTVQ GHV QLRIME NYCLMATKQK SNVEQALNTF TEIAASEKEH IPALLGMATA YMILKQTPRA RNQLKRIAKM NWNAIDAEEF EKSW LLLAD IYIQSAKYDM AEDLLKRCLR HNRSCCKAYE YMGYIMEKEQ AYTDAALNYE MAWKYSNRTN PAVGYKLAFN YLKAK RYVD SIDICHQVLE AHPTYPKIRK DILDKARASL RP |
-Macromolecule #5: Intraflagellar transport protein 140 homolog
| Macromolecule | Name: Intraflagellar transport protein 140 homolog / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 165.404281 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MALYYDHQIE APDAAGSPSF ISWHPVHPFL AVAYISTTST GSVDIYLEQG ECVPDTHVER PFRVASLCWH PTRLVLAVGW ETGEVTVFN KQDKEQHTMP LTHTADITVL RWSPSGNCLL SGDRLGVLLL WRLDQRGRVQ GTPLLKHEYG KHLTHCIFRL P PPGEDLVQ ...String: MALYYDHQIE APDAAGSPSF ISWHPVHPFL AVAYISTTST GSVDIYLEQG ECVPDTHVER PFRVASLCWH PTRLVLAVGW ETGEVTVFN KQDKEQHTMP LTHTADITVL RWSPSGNCLL SGDRLGVLLL WRLDQRGRVQ GTPLLKHEYG KHLTHCIFRL P PPGEDLVQ LAKAAVSGDE KALDMFNWKK SSSGSLLKMG SHEGLLFFVS LMDGTVHYVD EKGKTTQVVS ADSTIQMLFY ME KREALVV VTENLRLSLY TVPPEGKAEE VMKVKLSGKT GRRADIALIE GSLLVMAVGE AALRFWDIER GENYILSPDE KFG FEKGEN MNCVCYCKVK GLLAAGTDRG RVAMWRKVPD FLGSPGAEGK DRWALQTPTE LQGNITQIQW GSRKNLLAVN SVIS VAILS ERAMSSHFHQ QVAAMQVSPS LLNVCFLSTG VAHSLRTDMH ISGVFATKDA VAVWNGRQVA IFELSGAAIR SAGTF LCET PVLAMHEENV YTVESNRVQV RTWQGTVKQL LLFSETEGNP CFLDICGNFL VVGTDLAHFK SFDLSRREAK AHCSCR SLA ELVPGVGGIA SLRCSSSGST ISILPSKADN SPDSKICFYD VEMDTVTVFD FKTGQIDRRE TLSFNEQETN KSHLFVD EG LKNYVPVNHF WDQSEPRLFV CEAVQETPRS QPQSANGQPQ DGRAGPAADV LILSFFISEE HGFLLHESFP RPATSHSL L GMEVPYYYFT RKPEEADRED EVEPGCHHIP QMVSRRPLRD FVGLEDCDKA TRDAMLHFSF FVTIGDMDEA FKSIKLIKS EAVWENMARM CVKTQRLDVA KVCLGNMGHA RGARALREAE QEPELEARVA VLATQLGMLE DAEQLYRKCK RHDLLNKFYQ AAGRWQEAL QVAEHHDRVH LRSTYHRYAG HLEASADCSR ALSYYEKSDT HRFEVPRMLS EDLPSLELYV NKMKDKTLWR W WAQYLESQ GEMDAALHYY ELARDHFSLV RIHCFQGNVQ KAAQIANETG NLAASYHLAR QYESQEEVGQ AVHFYTRAQA FK NAIRLCK ENGLDDQLMN LALLSSPEDM IEAARYYEEK GVQMDRAVML YHKAGHFSKA LELAFATQQF VALQLIAEDL DET SDPALL ARCSDFFIEH SQYERAVELL LAARKYQEAL QLCLGQNMSI TEEMAEKMTV AKDSSDLPEE SRRELLEQIA DCCM RQGSY HLATKKYTQA GNKLKAMRAL LKSGDTEKIT FFASVSRQKE IYIMAANYLQ SLDWRKEPEI MKNIIGFYTK GRALD LLAG FYDACAQVEI DEYQNYDKAH GALTEAYKCL AKAKAKSPLD QETRLAQLQS RMALVKRFIQ ARRTYTEDPK ESIKQC ELL LEEPDLDSTI RIGDVYGFLV EHYVRKEEYQ TAYRFLEEMR RRLPLANMSY YVSPQAVDAV HRGLGLPLPR TVPEQVR HN SMEDARELDE EVVEEADDDP |
-Macromolecule #6: Intraflagellar transport protein 43 homolog
| Macromolecule | Name: Intraflagellar transport protein 43 homolog / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.55807 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEDLLDLDEE LRYSLATSRA KMGRRAQQES AQAENHLNGK NSSLTLTGET SSAKLPRCRQ GGWAGDSVKA SKFRRKASEE IEDFRLRPQ SLNGSDYGGD IPIIPDLEEV QEEDFVLQVA APPSIQIKRV MTYRDLDNDL MKYSAIQTLD GEIDLKLLTK V LAPEHEVR ...String: MEDLLDLDEE LRYSLATSRA KMGRRAQQES AQAENHLNGK NSSLTLTGET SSAKLPRCRQ GGWAGDSVKA SKFRRKASEE IEDFRLRPQ SLNGSDYGGD IPIIPDLEEV QEEDFVLQVA APPSIQIKRV MTYRDLDNDL MKYSAIQTLD GEIDLKLLTK V LAPEHEVR EDDVGWDWDH LFTEVSSEVL TEWDPLQTEK EDPAGQARHT |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 5 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 66.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: OTHER |
|---|---|
| Final angle assignment | Type: OTHER |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 67560 |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X