[English] 日本語
 Yorodumi
Yorodumi- EMDB-27587: Cryo-EM structure of the human Sec61 complex inhibited by ipomoea... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
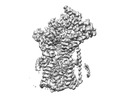
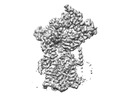
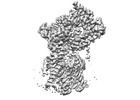
| Title | Cryo-EM structure of the human Sec61 complex inhibited by ipomoeassin F | |||||||||
 Map data Map data | Cryo-EM structure of the human Sec61 complex inhibited by ipomoeassin F | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  translocon / translocon /  inhibitor / inhibitor /  protein translocation / protein translocation /  PROTEIN TRANSPORT / PROTEIN TRANSPORT-INHIBITOR complex PROTEIN TRANSPORT / PROTEIN TRANSPORT-INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationendoplasmic reticulum Sec complex / endoplasmic reticulum quality control compartment / pronephric nephron development / cotranslational protein targeting to membrane / Ssh1 translocon complex / Sec61 translocon complex / protein targeting to ER / protein insertion into ER membrane /  post-translational protein targeting to endoplasmic reticulum membrane / SRP-dependent cotranslational protein targeting to membrane, translocation ...endoplasmic reticulum Sec complex / endoplasmic reticulum quality control compartment / pronephric nephron development / cotranslational protein targeting to membrane / Ssh1 translocon complex / Sec61 translocon complex / protein targeting to ER / protein insertion into ER membrane / post-translational protein targeting to endoplasmic reticulum membrane / SRP-dependent cotranslational protein targeting to membrane, translocation ...endoplasmic reticulum Sec complex / endoplasmic reticulum quality control compartment / pronephric nephron development / cotranslational protein targeting to membrane / Ssh1 translocon complex / Sec61 translocon complex / protein targeting to ER / protein insertion into ER membrane /  post-translational protein targeting to endoplasmic reticulum membrane / SRP-dependent cotranslational protein targeting to membrane, translocation / post-translational protein targeting to endoplasmic reticulum membrane / SRP-dependent cotranslational protein targeting to membrane, translocation /  signal sequence binding / SRP-dependent cotranslational protein targeting to membrane / signal sequence binding / SRP-dependent cotranslational protein targeting to membrane /  post-translational protein targeting to membrane, translocation / endoplasmic reticulum organization / retrograde protein transport, ER to cytosol / post-translational protein targeting to membrane, translocation / endoplasmic reticulum organization / retrograde protein transport, ER to cytosol /  epidermal growth factor binding / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / SRP-dependent cotranslational protein targeting to membrane / protein transmembrane transporter activity / : / epidermal growth factor binding / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / SRP-dependent cotranslational protein targeting to membrane / protein transmembrane transporter activity / : /  calcium channel activity / calcium channel activity /  ribosome binding / ER-Phagosome pathway / endoplasmic reticulum membrane / ribosome binding / ER-Phagosome pathway / endoplasmic reticulum membrane /  endoplasmic reticulum / endoplasmic reticulum /  RNA binding / RNA binding /  membrane / membrane /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.01 Å cryo EM / Resolution: 3.01 Å | |||||||||
 Authors Authors | Park E / Itskanov S | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: A common mechanism of Sec61 translocon inhibition by small molecules. Authors: Samuel Itskanov / Laurie Wang / Tina Junne / Rumi Sherriff / Li Xiao / Nicolas Blanchard / Wei Q Shi / Craig Forsyth / Dominic Hoepfner / Martin Spiess / Eunyong Park /    Abstract: The Sec61 complex forms a protein-conducting channel in the endoplasmic reticulum membrane that is required for secretion of soluble proteins and production of many membrane proteins. Several natural ...The Sec61 complex forms a protein-conducting channel in the endoplasmic reticulum membrane that is required for secretion of soluble proteins and production of many membrane proteins. Several natural and synthetic small molecules specifically inhibit Sec61, generating cellular effects that are useful for therapeutic purposes, but their inhibitory mechanisms remain unclear. Here we present near-atomic-resolution structures of human Sec61 inhibited by a comprehensive panel of structurally distinct small molecules-cotransin, decatransin, apratoxin, ipomoeassin, mycolactone, cyclotriazadisulfonamide and eeyarestatin. All inhibitors bind to a common lipid-exposed pocket formed by the partially open lateral gate and plug domain of Sec61. Mutations conferring resistance to the inhibitors are clustered at this binding pocket. The structures indicate that Sec61 inhibitors stabilize the plug domain in a closed state, thereby preventing the protein-translocation pore from opening. Our study provides the atomic details of Sec61-inhibitor interactions and the structural framework for further pharmacological studies and drug design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27587.map.gz emd_27587.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27587-v30.xml emd-27587-v30.xml emd-27587.xml emd-27587.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27587_fsc.xml emd_27587_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_27587.png emd_27587.png | 92.4 KB | ||
| Others |  emd_27587_half_map_1.map.gz emd_27587_half_map_1.map.gz emd_27587_half_map_2.map.gz emd_27587_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27587 http://ftp.pdbj.org/pub/emdb/structures/EMD-27587 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27587 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27587 | HTTPS FTP |
-Related structure data
| Related structure data |  8do1MC  8dnvC  8dnwC  8dnxC  8dnyC  8dnzC  8do0C  8do2C  8do3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27587.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27587.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the human Sec61 complex inhibited by ipomoeassin F | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of the human Sec61 complex inhibited...
| File | emd_27587_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the human Sec61 complex inhibited by ipomoeassin F | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM structure of the human Sec61 complex inhibited...
| File | emd_27587_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the human Sec61 complex inhibited by ipomoeassin F | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : A human-yeast chimeric Sec complex treated with ipomoeassin F
| Entire | Name: A human-yeast chimeric Sec complex treated with ipomoeassin F |
|---|---|
| Components |
|
-Supramolecule #1: A human-yeast chimeric Sec complex treated with ipomoeassin F
| Supramolecule | Name: A human-yeast chimeric Sec complex treated with ipomoeassin F type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / Organelle: endoplasmic reticulum Homo sapiens (human) / Organelle: endoplasmic reticulum |
-Macromolecule #1: Protein transport protein Sec61 subunit alpha isoform 1
| Macromolecule | Name: Protein transport protein Sec61 subunit alpha isoform 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.202438 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MAIKFLEVIK PFCVILPEIQ KPERKIQFKE KVLWTAITLF IFLVCCQIPL FGIMSSDSAD PFYWMRVILA SNRGTLMELG ISPIVTSGL IMQLLAGAKI IEVGDTPKDR ALFNGAQKLF GMIITIGQSI VYVMTGMYGD PSEMGAGICL LITIQLFVAG L IVLLLDEL ...String: MAIKFLEVIK PFCVILPEIQ KPERKIQFKE KVLWTAITLF IFLVCCQIPL FGIMSSDSAD PFYWMRVILA SNRGTLMELG ISPIVTSGL IMQLLAGAKI IEVGDTPKDR ALFNGAQKLF GMIITIGQSI VYVMTGMYGD PSEMGAGICL LITIQLFVAG L IVLLLDEL LQKGYGLGSG ISLFIATNIC ETIVWKAFSP TTVNTGRGME FEGAIIALFH LLATRTDKVR ALREAFYRQN LP NLMNLIA TIFVFAVVIY FQGFRYELPI RSTKVRGQIG IYPIKLFYTS NIPIILQSAL VSNLYVISQM LSARFSGNLL VSL LGTWSD TSSGGPARAY PVGGLCYYLS PPESFGSVLE DPVHAVVYIV FMLGSCAFFS KTWIEVSGSS PRDIAKQFKD QGMV INGKR ETSIYRELKK IIPTAAAFGG LCIGALSVLA DFLGAIGSGT GILLAVTIIY QYFEIFVKEQ SEVGSMGALL F UniProtKB:  Protein transport protein Sec61 subunit alpha isoform 1 Protein transport protein Sec61 subunit alpha isoform 1 |
-Macromolecule #2: Protein transport protein Sec61 subunit gamma
| Macromolecule | Name: Protein transport protein Sec61 subunit gamma / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.752325 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MDQVMQFVEP SRQFVKDSIR LVKRCTKPDR KEFQKIAMAT AIGFAIMGFI GFFVKLIHIP INNIIVGG UniProtKB:  Protein transport protein Sec61 subunit gamma Protein transport protein Sec61 subunit gamma |
-Macromolecule #3: Protein transport protein Sec61 subunit beta
| Macromolecule | Name: Protein transport protein Sec61 subunit beta / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.987456 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MPGPTPSGTN VGSSGRSPSK AVAARAAGST VRQRKNASCG TRSAGRTTSA GTGGMWRFYT EDSPGLKVGP VPVLVMSLLF IASVFMLHI WGKYTRS UniProtKB:  Protein transport protein Sec61 subunit beta Protein transport protein Sec61 subunit beta |
-Macromolecule #4: [(1~{S},3~{R},4~{S},5~{R},6~{R},8~{R},10~{S},23~{R},24~{R},25~{R}...
| Macromolecule | Name: [(1~{S},3~{R},4~{S},5~{R},6~{R},8~{R},10~{S},23~{R},24~{R},25~{R},26~{R})-5-acetyloxy-6-methyl-4,26-bis(oxidanyl)-17,20-bis(oxidanylidene)-10-pentyl-24-[(~{E})-3-phenylprop-2-enoyl]oxy- ...Name: [(1~{S},3~{R},4~{S},5~{R},6~{R},8~{R},10~{S},23~{R},24~{R},25~{R},26~{R})-5-acetyloxy-6-methyl-4,26-bis(oxidanyl)-17,20-bis(oxidanylidene)-10-pentyl-24-[(~{E})-3-phenylprop-2-enoyl]oxy-2,7,9,21,27-pentaoxatricyclo[21.3.1.0^{3,8}]heptacosan-25-yl] (~{E})-2-methylbut-2-enoate type: ligand / ID: 4 / Number of copies: 1 / Formula: SXF |
|---|---|
| Molecular weight | Theoretical: 830.954 Da |
| Chemical component information | 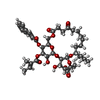 ChemComp-SXF: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 4 seconds before plunging. | ||||||||||||||||||
| Details | Reconsitituted into a peptidisc. Monodisperse peak from a Superose 6 column. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8do1: |
 Movie
Movie Controller
Controller





















 Z
Z Y
Y X
X


















