[English] 日本語
 Yorodumi
Yorodumi- EMDB-27248: Sub-tomogram averaged map of full-length post-fusion CHIKV E1 gly... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
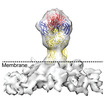
| Title | Sub-tomogram averaged map of full-length post-fusion CHIKV E1 glycoprotein trimer | |||||||||
 Map data Map data | Sub-tomogram averaged map of membrane attached full-length post-fusion CHIKV E1 glycoprotein trimer | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information togavirin / T=4 icosahedral viral capsid / togavirin / T=4 icosahedral viral capsid /  virion assembly / virion assembly /  small molecule binding / host cell endosome / symbiont-mediated suppression of host toll-like receptor signaling pathway / clathrin-dependent endocytosis of virus by host cell / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / small molecule binding / host cell endosome / symbiont-mediated suppression of host toll-like receptor signaling pathway / clathrin-dependent endocytosis of virus by host cell / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane /  viral envelope ... viral envelope ... togavirin / T=4 icosahedral viral capsid / togavirin / T=4 icosahedral viral capsid /  virion assembly / virion assembly /  small molecule binding / host cell endosome / symbiont-mediated suppression of host toll-like receptor signaling pathway / clathrin-dependent endocytosis of virus by host cell / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / small molecule binding / host cell endosome / symbiont-mediated suppression of host toll-like receptor signaling pathway / clathrin-dependent endocytosis of virus by host cell / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane /  viral envelope / host cell nucleus / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / viral envelope / host cell nucleus / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity /  proteolysis / proteolysis /  RNA binding / RNA binding /  membrane membraneSimilarity search - Function | |||||||||
| Biological species |   Chikungunya virus strain S27-African prototype Chikungunya virus strain S27-African prototype | |||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 27.2 Å cryo EM / Resolution: 27.2 Å | |||||||||
 Authors Authors | Mangala Prasad V / Lee KK | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Visualization of conformational changes and membrane remodeling leading to genome delivery by viral class-II fusion machinery. Authors: Vidya Mangala Prasad / Jelle S Blijleven / Jolanda M Smit / Kelly K Lee /    Abstract: Chikungunya virus (CHIKV) is a human pathogen that delivers its genome to the host cell cytoplasm through endocytic low pH-activated membrane fusion mediated by class-II fusion proteins. Though ...Chikungunya virus (CHIKV) is a human pathogen that delivers its genome to the host cell cytoplasm through endocytic low pH-activated membrane fusion mediated by class-II fusion proteins. Though structures of prefusion, icosahedral CHIKV are available, structural characterization of virion interaction with membranes has been limited. Here, we have used cryo-electron tomography to visualize CHIKV's complete membrane fusion pathway, identifying key intermediary glycoprotein conformations coupled to membrane remodeling events. Using sub-tomogram averaging, we elucidate features of the low pH-exposed virion, nucleocapsid and full-length E1-glycoprotein's post-fusion structure. Contrary to class-I fusion systems, CHIKV achieves membrane apposition by protrusion of extended E1-glycoprotein homotrimers into the target membrane. The fusion process also features a large hemifusion diaphragm that transitions to a wide pore for intact nucleocapsid delivery. Our analyses provide comprehensive ultrastructural insights into the class-II virus fusion system function and direct mechanistic characterization of the fundamental process of protein-mediated membrane fusion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27248.map.gz emd_27248.map.gz | 232.3 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27248-v30.xml emd-27248-v30.xml emd-27248.xml emd-27248.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27248_fsc.xml emd_27248_fsc.xml | 4.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_27248.png emd_27248.png | 106.7 KB | ||
| Others |  emd_27248_half_map_1.map.gz emd_27248_half_map_1.map.gz emd_27248_half_map_2.map.gz emd_27248_half_map_2.map.gz | 232.1 KB 232 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27248 http://ftp.pdbj.org/pub/emdb/structures/EMD-27248 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27248 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27248 | HTTPS FTP |
-Related structure data
| Related structure data |  8d87MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27248.map.gz / Format: CCP4 / Size: 251 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27248.map.gz / Format: CCP4 / Size: 251 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sub-tomogram averaged map of membrane attached full-length post-fusion CHIKV E1 glycoprotein trimer | ||||||||||||||||||||
| Voxel size | X=Y=Z: 6.612 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Odd-half map
| File | emd_27248_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Odd-half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
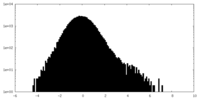
| Density Histograms |
-Half map: Even half map
| File | emd_27248_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Even half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Sub-tomogram averaged map of post-fusion E1 glycoprotein trimer f...
| Entire | Name: Sub-tomogram averaged map of post-fusion E1 glycoprotein trimer from Chikungunya virus |
|---|---|
| Components |
|
-Supramolecule #1: Sub-tomogram averaged map of post-fusion E1 glycoprotein trimer f...
| Supramolecule | Name: Sub-tomogram averaged map of post-fusion E1 glycoprotein trimer from Chikungunya virus type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Chikungunya virus strain S27-African prototype Chikungunya virus strain S27-African prototype |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Spike glycoprotein E1
| Macromolecule | Name: Spike glycoprotein E1 / type: protein_or_peptide / ID: 1 Details: The previously determined X-ray crystal structure PDB ID 1RER was rigidly docked into the sub-tomogram averaged map but no further refinement was performed Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Chikungunya virus strain S27-African prototype Chikungunya virus strain S27-African prototype |
| Molecular weight | Theoretical: 42.690125 KDa |
| Sequence | String: YEHSTVMPNV VGFPYKAHIE RPGYSPLTLQ MQVVETSLEP TLNLEYITCE YKTVVPSPYV KCCGASECST KEKPDYQCKV YTGVYPFMW GGAYCFCDSE NTQLSEAYVD RSDVCRHDHA SAYKAHTASL KAKVRVMYGN VNQTVDVYVN GDHAVTIGGT Q FIFGPLSS ...String: YEHSTVMPNV VGFPYKAHIE RPGYSPLTLQ MQVVETSLEP TLNLEYITCE YKTVVPSPYV KCCGASECST KEKPDYQCKV YTGVYPFMW GGAYCFCDSE NTQLSEAYVD RSDVCRHDHA SAYKAHTASL KAKVRVMYGN VNQTVDVYVN GDHAVTIGGT Q FIFGPLSS AWTPFDNKIV VYKDEVFNQD FPPYGSGQPG RFGDIQSRTV ESNDLYANTA LKLARPSPGM VHVPYTQTPS GF KYWLKEK GTALNTKAPF GCQIKTNPVR AMNCAVGNIP VSMNLPDSAF TRIVEAPTII DLTCTVATCT HSSDFGGVLT LTY KTNKNG DCSVHSHSNV ATLQEATAKV KTAGKVTLHF STASASPSFV VSLCSARATC SASCEPPKDH IVPYA |
-Macromolecule #5: BROMIDE ION
| Macromolecule | Name: BROMIDE ION / type: ligand / ID: 5 / Number of copies: 3 / Formula: BR |
|---|---|
| Molecular weight | Theoretical: 79.904 Da |
| Chemical component information |  ChemComp-BR: |
-Macromolecule #6: HOLMIUM ATOM
| Macromolecule | Name: HOLMIUM ATOM / type: ligand / ID: 6 / Number of copies: 4 / Formula: HO |
|---|---|
| Molecular weight | Theoretical: 164.93 Da |
| Chemical component information |  ChemComp-HO: |
-Macromolecule #7: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 139 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 5.1 / Details: Hepes Buffer Saline |
|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 7-8 seconds. |
| Details | Sub-volumes picked from liposome membrane surface |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 53000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 53000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 70.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X