+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of GSK682753A-bound EBI2/GPR183 | ||||||||||||
 Map data Map data | GSK682753A-bound EBI2/GPR183 | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of astrocyte chemotaxis / B cell activation involved in immune response / dendritic cell homeostasis / T follicular helper cell differentiation / mature B cell differentiation involved in immune response / Class A/1 (Rhodopsin-like receptors) / T cell chemotaxis /  oxysterol binding / leukocyte chemotaxis / dendritic cell chemotaxis ...regulation of astrocyte chemotaxis / B cell activation involved in immune response / dendritic cell homeostasis / T follicular helper cell differentiation / mature B cell differentiation involved in immune response / Class A/1 (Rhodopsin-like receptors) / T cell chemotaxis / oxysterol binding / leukocyte chemotaxis / dendritic cell chemotaxis ...regulation of astrocyte chemotaxis / B cell activation involved in immune response / dendritic cell homeostasis / T follicular helper cell differentiation / mature B cell differentiation involved in immune response / Class A/1 (Rhodopsin-like receptors) / T cell chemotaxis /  oxysterol binding / leukocyte chemotaxis / dendritic cell chemotaxis / plasma membrane => GO:0005886 / oxysterol binding / leukocyte chemotaxis / dendritic cell chemotaxis / plasma membrane => GO:0005886 /  humoral immune response / positive regulation of B cell proliferation / osteoclast differentiation / G protein-coupled receptor activity / G alpha (i) signalling events / humoral immune response / positive regulation of B cell proliferation / osteoclast differentiation / G protein-coupled receptor activity / G alpha (i) signalling events /  adaptive immune response / positive regulation of ERK1 and ERK2 cascade / adaptive immune response / positive regulation of ERK1 and ERK2 cascade /  immune response / G protein-coupled receptor signaling pathway / immune response / G protein-coupled receptor signaling pathway /  plasma membrane plasma membraneSimilarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.98 Å cryo EM / Resolution: 2.98 Å | ||||||||||||
 Authors Authors | Chen H / Huang W / Li X | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response. Authors: Hongwen Chen / Weijiao Huang / Xiaochun Li /  Abstract: Oxysterols induce the migration of B-lymphocytes and dendritic cells to interfollicular regions of lymphoid tissues through binding the EBI2 (GPR183) to stimulate effective adaptive immunity and ...Oxysterols induce the migration of B-lymphocytes and dendritic cells to interfollicular regions of lymphoid tissues through binding the EBI2 (GPR183) to stimulate effective adaptive immunity and antibody production during infection. Aberrant EBI2 signaling is implicated in inflammatory bowel disease, sclerosis, and infectious disease. Here, we report the cryo-EM structures of an EBI2-G signaling complex with its endogenous agonist 7α,25-OHC and that of an inactive EBI2 bound to the inverse agonist GSK682753A. These structures reveal an agonist binding site for the oxysterol and a potential ligand entrance site exposed to the lipid bilayer. Mutations within the oxysterol binding site and the Gα interface attenuate G protein signaling and abolish oxysterol-mediated cell migration indicating that G protein signaling directly involves in the oxysterol-EBI2 pathway. Together, these findings provide new insight into how EBI2 is activated by an oxysterol ligand and will facilitate the development of therapeutic approaches that target EBI2-linked diseases. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26135.map.gz emd_26135.map.gz | 53.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26135-v30.xml emd-26135-v30.xml emd-26135.xml emd-26135.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26135.png emd_26135.png | 70 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26135 http://ftp.pdbj.org/pub/emdb/structures/EMD-26135 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26135 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26135 | HTTPS FTP |
-Related structure data
| Related structure data |  7tuyMC  7tuzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26135.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26135.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GSK682753A-bound EBI2/GPR183 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : complex of GSK682753A-bound EBI2/GPR183-BRIL chimera with anti-BR...
| Entire | Name: complex of GSK682753A-bound EBI2/GPR183-BRIL chimera with anti-BRIL Fab and anti-Fab nanobody |
|---|---|
| Components |
|
-Supramolecule #1: complex of GSK682753A-bound EBI2/GPR183-BRIL chimera with anti-BR...
| Supramolecule | Name: complex of GSK682753A-bound EBI2/GPR183-BRIL chimera with anti-BRIL Fab and anti-Fab nanobody type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain
| Supramolecule | Name: anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1, #3 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
-Supramolecule #3: anti-Fab Nanobody
| Supramolecule | Name: anti-Fab Nanobody / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
-Supramolecule #4: G-protein coupled receptor 183,Soluble cytochrome b562 fusion
| Supramolecule | Name: G-protein coupled receptor 183,Soluble cytochrome b562 fusion type: complex / Chimera: Yes / ID: 4 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: anti-BRIL Fab Heavy chain
| Macromolecule | Name: anti-BRIL Fab Heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.321084 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVVDFSLHWV RQAPGKGLEW VAYISSSSGS TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARWGYWPGEP WWKAFDYWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVVDFSLHWV RQAPGKGLEW VAYISSSSGS TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARWGYWPGEP WWKAFDYWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK S |
-Macromolecule #2: anti-Fab Nanobody
| Macromolecule | Name: anti-Fab Nanobody / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.071431 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: HHHHHHGENL YFQGSQVQLQ ESGGGLVQPG GSLRLSCAAS GRTISRYAMS WFRQAPGKER EFVAVARRSG DGAFYADSVQ GRFTVSRDD AKNTVYLQMN SLKPEDTAVY YCAIDSDTFY SGSYDYWGQG TQVTVSS |
-Macromolecule #3: anti-BRIL Fab Light chain
| Macromolecule | Name: anti-BRIL Fab Light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.483062 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYLYYSLVT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYLYYSLVT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGE |
-Macromolecule #4: G-protein coupled receptor 183,Soluble cytochrome b562 fusion
| Macromolecule | Name: G-protein coupled receptor 183,Soluble cytochrome b562 fusion type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.364539 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDIQMANNFT PPSATPQGND CDLYAHHSTA RIVMPLHYSL VFIIGLVGNL LALVVIVQNR KKINSTTLYS TNLVISDILF TTALPTRIA YYAMGFDWRI GDALCRITAL VFYINTYAGV NFMTCLSIDR FIAVVHPLRY NKIKRIEHAK GVCIFVWILV F AQTLPLLI ...String: MDIQMANNFT PPSATPQGND CDLYAHHSTA RIVMPLHYSL VFIIGLVGNL LALVVIVQNR KKINSTTLYS TNLVISDILF TTALPTRIA YYAMGFDWRI GDALCRITAL VFYINTYAGV NFMTCLSIDR FIAVVHPLRY NKIKRIEHAK GVCIFVWILV F AQTLPLLI NPMSKQEAER ITCMEYPNFE ETKSLPWILL GACFIGYVLP LIIILICYSQ ICCKLFRARR QLADLEDNWE TL NDNLKVI EKADNAAQVK DALTKMRAAA LDAQKATPPK LEDKSPDSPE MKDFRHGFDI LVGQIDDALK LANEGKVKEA QAA AEQLKT TRNAYIQKYL ERARSTLSGV NKKALNTIIL IIVVFVLCFT PYHVAIIQHM IKKLRFSNFL ECSQRHSFQI SLHF TVCLM NFNCCMDPFI YFFACKGYKR KVMRMLKRQV SVSISSAVKS APEENSREMT ETQMMIHSKS SNGKDYKDDD DK |
-Macromolecule #5: 8-[(2E)-3-(4-chlorophenyl)prop-2-enoyl]-3-[(3,4-dichlorophenyl)me...
| Macromolecule | Name: 8-[(2E)-3-(4-chlorophenyl)prop-2-enoyl]-3-[(3,4-dichlorophenyl)methyl]-1-oxa-3,8-diazaspiro[4.5]decan-2-one type: ligand / ID: 5 / Number of copies: 1 / Formula: KKF |
|---|---|
| Molecular weight | Theoretical: 479.783 Da |
| Chemical component information | 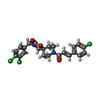 ChemComp-KKF: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.98 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 222283 |
 Movie
Movie Controller
Controller














