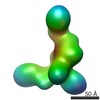
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25708 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GPC2 HEP CT3 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of protein localization to membrane / Defective B3GALT6 causes EDSP2 and SEMDJL1 / Defective B4GALT7 causes EDS, progeroid type / Defective B3GAT3 causes JDSSDHD / Defective EXT2 causes exostoses 2 / Defective EXT1 causes exostoses 1, TRPS2 and CHDS / A tetrasaccharide linker sequence is required for GAG synthesis / HS-GAG biosynthesis / HS-GAG degradation / smoothened signaling pathway ...regulation of protein localization to membrane / Defective B3GALT6 causes EDSP2 and SEMDJL1 / Defective B4GALT7 causes EDS, progeroid type / Defective B3GAT3 causes JDSSDHD / Defective EXT2 causes exostoses 2 / Defective EXT1 causes exostoses 1, TRPS2 and CHDS / A tetrasaccharide linker sequence is required for GAG synthesis / HS-GAG biosynthesis / HS-GAG degradation / smoothened signaling pathway /  regulation of signal transduction / Retinoid metabolism and transport / lysosomal lumen / positive regulation of neuron projection development / neuron differentiation / Golgi lumen / regulation of signal transduction / Retinoid metabolism and transport / lysosomal lumen / positive regulation of neuron projection development / neuron differentiation / Golgi lumen /  cell migration / collagen-containing extracellular matrix / Attachment and Entry / cell migration / collagen-containing extracellular matrix / Attachment and Entry /  synapse / synapse /  cell surface / cell surface /  endoplasmic reticulum / endoplasmic reticulum /  extracellular space / extracellular space /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 21.0 Å negative staining / Resolution: 21.0 Å | |||||||||
 Authors Authors | Zhu J / Cachau R / De Val Alda N / Li N / Ho M | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep Med / Year: 2021 Journal: Cell Rep Med / Year: 2021Title: CAR T cells targeting tumor-associated exons of glypican 2 regress neuroblastoma in mice. Authors: Nan Li / Madeline B Torres / Madeline R Spetz / Ruixue Wang / Luyi Peng / Meijie Tian / Christopher M Dower / Rosa Nguyen / Ming Sun / Chin-Hsien Tai / Natalia de Val / Raul Cachau / Xiaolin ...Authors: Nan Li / Madeline B Torres / Madeline R Spetz / Ruixue Wang / Luyi Peng / Meijie Tian / Christopher M Dower / Rosa Nguyen / Ming Sun / Chin-Hsien Tai / Natalia de Val / Raul Cachau / Xiaolin Wu / Stephen M Hewitt / Rosandra N Kaplan / Javed Khan / Brad St Croix / Carol J Thiele / Mitchell Ho /  Abstract: Targeting solid tumors must overcome several major obstacles, in particular, the identification of elusive tumor-specific antigens. Here, we devise a strategy to help identify tumor-specific epitopes. ...Targeting solid tumors must overcome several major obstacles, in particular, the identification of elusive tumor-specific antigens. Here, we devise a strategy to help identify tumor-specific epitopes. Glypican 2 (GPC2) is overexpressed in neuroblastoma. Using RNA sequencing (RNA-seq) analysis, we show that exon 3 and exons 7-10 of GPC2 are expressed in cancer but are minimally expressed in normal tissues. Accordingly, we discover a monoclonal antibody (CT3) that binds exons 3 and 10 and visualize the complex structure of CT3 and GPC2 by electron microscopy. The potential of this approach is exemplified by designing CT3-derived chimeric antigen receptor (CAR) T cells that regress neuroblastoma in mice. Genomic sequencing of T cells recovered from mice reveals the CAR integration sites that may contribute to CAR T cell proliferation and persistence. These studies demonstrate how RNA-seq data can be exploited to help identify tumor-associated exons that can be targeted by CAR T cell therapies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25708.map.gz emd_25708.map.gz | 23.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25708-v30.xml emd-25708-v30.xml emd-25708.xml emd-25708.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25708_fsc.xml emd_25708_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_25708.png emd_25708.png | 21.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25708 http://ftp.pdbj.org/pub/emdb/structures/EMD-25708 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25708 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25708 | HTTPS FTP |
-Related structure data
| Related structure data |  7t62MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25708.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25708.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 2.19 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : GPC2 HET CT3 complex
| Entire | Name: GPC2 HET CT3 complex |
|---|---|
| Components |
|
-Supramolecule #1: GPC2 HET CT3 complex
| Supramolecule | Name: GPC2 HET CT3 complex / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Macromolecule #1: Glypican-2
| Macromolecule | Name: Glypican-2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.002066 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPGPGSEAKV TRSCAETRQV LGARGYSLNL IPPALISGEH LRVCPQEYTC CSSETEQRLI RETEATFRGL VEDSGSFLVH TLAARHRKF DEFFLEMLSV AQHSLTQLFS HSYGRLYAQH ALIFNGLFSR LRDFYGESGE GLDDTLADFW AQLLERVFPL L HPQYSFPP ...String: GPGPGSEAKV TRSCAETRQV LGARGYSLNL IPPALISGEH LRVCPQEYTC CSSETEQRLI RETEATFRGL VEDSGSFLVH TLAARHRKF DEFFLEMLSV AQHSLTQLFS HSYGRLYAQH ALIFNGLFSR LRDFYGESGE GLDDTLADFW AQLLERVFPL L HPQYSFPP DYLLCLSRLA SSTDGSLQPF GDSPRRLRLQ ITRTLVAARA FVQGLETGRN VVSEALKVPV SEGCSQALMR LI GCPLCRG VPSLMPCQGF CLNVVRGCLS SRGLEPDWGN YLDGLLILAD KLQGPFSFEL TAESIGVKIS EGLMYLQENS AKV SAQVFQ ECGPPDPVPA RNRRAPPPRE EAGRLWSMVT EEERPTTAAG TNLHRLVWEL RERLARMRGF WARLSLTVCG DSRM AADAS LEAAPCWTGA GRGRYLPPVV GGSPAEQVNN PELKVDASGP DVPTRRRRLQ LRAATARMKT AALGHDLDGQ DADED ASGS GGGQQYADDW MAGAVAPPAR PPRPPYPPRR DGSGGKGGGG SARYNQGRSR SGGASIGFHT QTILILSLSA LALLGP R |
-Macromolecule #2: CT3
| Macromolecule | Name: CT3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 47.359551 KDa |
| Recombinant expression | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Sequence | String: EVQLQQSGPE LVKPGASVKM SCKASRFTFT DYNIHWVKQS PGKTLEWIGY INPNNGDIFY KQKFNGKATL TINKSSNTAY MELRSLTSE DSAVYYCVRS SNIRYTFDRF FDVWGTGTTV TVSSAKTTPP SVYPLAPGSA AQTNSMVTLG CLVKGYFPEP V TVTWNSGS ...String: EVQLQQSGPE LVKPGASVKM SCKASRFTFT DYNIHWVKQS PGKTLEWIGY INPNNGDIFY KQKFNGKATL TINKSSNTAY MELRSLTSE DSAVYYCVRS SNIRYTFDRF FDVWGTGTTV TVSSAKTTPP SVYPLAPGSA AQTNSMVTLG CLVKGYFPEP V TVTWNSGS LSSGVHTFPA VLESDLYTLS SSVTVPSSPR PSETVTCNVA HPASSTKVDK KIENVLTQSP AIMSASLGEK VT MSCRASS SVNYIYWYQQ KSDASPKLWI YYTSNLAPGV PARFSGSGSG NSYSLTISSM EGEDAATYYC QQFSSSPSTF GTG TKLELK RADAAPTVSI FPPSSEQLTS GGASVVCFLN NFYPKDINVK WKIDGSERQN GVLNSWTDQD SKDSTYSMSS TLTL TKDEY ERHNSYTCEA THKTSTSPIV KSFNRNEC |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Staining | Type: NEGATIVE / Material: uranyl formate Details: A 3 uL aliquot containing ~0.01 mg/mL of the samples was applied for 20 s onto a carbon-coated 200 Cu mesh grid (Electron Microscopy Sciences, Protochips, Inc.) that had been glow discharged ...Details: A 3 uL aliquot containing ~0.01 mg/mL of the samples was applied for 20 s onto a carbon-coated 200 Cu mesh grid (Electron Microscopy Sciences, Protochips, Inc.) that had been glow discharged at 30 mA for 30 s (Pelco easiGlow, Ted Pella, Inc.), then negatively stained with 0.7% (w/v) uranyl formate for 40 sec. |
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 100000 Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 100000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI EAGLE (2k x 2k) / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Number real images: 200 / Average electron dose: 40.0 e/Å2 |
 Movie
Movie Controller
Controller