[English] 日本語
 Yorodumi
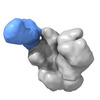
Yorodumi- EMDB-25676: The Envelope Glycoprotein SIVmac239.K180S SOSIP trimer in complex... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 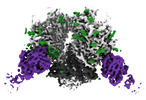 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Envelope Glycoprotein SIVmac239.K180S SOSIP trimer in complex with 3 copies of the neutralizing antibody K11 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SIV / Env /  vaccine / vaccine /  neutralizing antibody / neutralizing antibody /  glycoprotein / glycoprotein /  VIRAL PROTEIN-IMMUNE SYSTEM complex VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane fusion involved in viral entry into host cell / host cell endosome membrane / membrane => GO:0016020 / symbiont entry into host cell /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Simian immunodeficiency virus / Simian immunodeficiency virus /   Macaca mulatta (Rhesus monkey) Macaca mulatta (Rhesus monkey) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Berndsen ZT / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Molecular insights into antibody-mediated protection against the prototypic simian immunodeficiency virus. Authors: Fangzhu Zhao / Zachary T Berndsen / Nuria Pedreño-Lopez / Alison Burns / Joel D Allen / Shawn Barman / Wen-Hsin Lee / Srirupa Chakraborty / Sandrasegaram Gnanakaran / Leigh M Sewall / ...Authors: Fangzhu Zhao / Zachary T Berndsen / Nuria Pedreño-Lopez / Alison Burns / Joel D Allen / Shawn Barman / Wen-Hsin Lee / Srirupa Chakraborty / Sandrasegaram Gnanakaran / Leigh M Sewall / Gabriel Ozorowski / Oliver Limbo / Ge Song / Peter Yong / Sean Callaghan / Jessica Coppola / Kim L Weisgrau / Jeffrey D Lifson / Rebecca Nedellec / Thomas B Voigt / Fernanda Laurino / Johan Louw / Brandon C Rosen / Michael Ricciardi / Max Crispin / Ronald C Desrosiers / Eva G Rakasz / David I Watkins / Raiees Andrabi / Andrew B Ward / Dennis R Burton / Devin Sok /   Abstract: SIVmac239 infection of macaques is a favored model of human HIV infection. However, the SIVmac239 envelope (Env) trimer structure, glycan occupancy, and the targets and ability of neutralizing ...SIVmac239 infection of macaques is a favored model of human HIV infection. However, the SIVmac239 envelope (Env) trimer structure, glycan occupancy, and the targets and ability of neutralizing antibodies (nAbs) to protect against SIVmac239 remain unknown. Here, we report the isolation of SIVmac239 nAbs that recognize a glycan hole and the V1/V4 loop. A high-resolution structure of a SIVmac239 Env trimer-nAb complex shows many similarities to HIV and SIVcpz Envs, but with distinct V4 features and an extended V1 loop. Moreover, SIVmac239 Env has a higher glycan shield density than HIV Env that may contribute to poor or delayed nAb responses in SIVmac239-infected macaques. Passive transfer of a nAb protects macaques from repeated intravenous SIVmac239 challenge at serum titers comparable to those described for protection of humans against HIV infection. Our results provide structural insights for vaccine design and shed light on antibody-mediated protection in the SIV model. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25676.map.gz emd_25676.map.gz | 97.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25676-v30.xml emd-25676-v30.xml emd-25676.xml emd-25676.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25676_fsc.xml emd_25676_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_25676.png emd_25676.png | 98.9 KB | ||
| Masks |  emd_25676_msk_1.map emd_25676_msk_1.map | 103 MB |  Mask map Mask map | |
| Others |  emd_25676_half_map_1.map.gz emd_25676_half_map_1.map.gz emd_25676_half_map_2.map.gz emd_25676_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25676 http://ftp.pdbj.org/pub/emdb/structures/EMD-25676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25676 | HTTPS FTP |
-Related structure data
| Related structure data |  7t4gMC  7t2pC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25676.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25676.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25676_msk_1.map emd_25676_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25676_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_25676_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SIVmac239.K180S SOSIP trimer in complex with 3 copies of K11 IgG
| Entire | Name: SIVmac239.K180S SOSIP trimer in complex with 3 copies of K11 IgG |
|---|---|
| Components |
|
-Supramolecule #1: SIVmac239.K180S SOSIP trimer in complex with 3 copies of K11 IgG
| Supramolecule | Name: SIVmac239.K180S SOSIP trimer in complex with 3 copies of K11 IgG type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Simian immunodeficiency virus Simian immunodeficiency virus |
-Macromolecule #1: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 60.324441 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGCLGNQLLI AILLLSVYGI YCTLYVTVFY GVPAWRNATI PLFCATKNRD TWGTTQCLPD NGDYSEMALN VTESFDAWNN TVTEQAIED VWQLFETSIK PCVKLSPLCI TMRCNKSETD RWGLTKSITT TASTTSTTAS AKVDMVNETS SCIAQDNCTG L EQEQMISC ...String: MGCLGNQLLI AILLLSVYGI YCTLYVTVFY GVPAWRNATI PLFCATKNRD TWGTTQCLPD NGDYSEMALN VTESFDAWNN TVTEQAIED VWQLFETSIK PCVKLSPLCI TMRCNKSETD RWGLTKSITT TASTTSTTAS AKVDMVNETS SCIAQDNCTG L EQEQMISC KFNMTGLKRD KSKEYNETWY SADLVCEQGN NTGNESRCYM NHCNTSVIQE SCDKHYWDAI RFRYCAPPGY AL LRCNDTN YSGFMPKCSK VVVSSCTRMM ETQTSTWFGF NGTRAENRTY IYWHGRDNRT IISLNKYYNL TMKCRRPGNK TVL PVTIMS GLVFHSQPIN DRPKQAWCWF GGKWKDAIKE VKQTIVKHPR YTGTNNTDKI NLTAPGGGDP EVTFMWTNCR GEFL YCKMN WFLNWVEDRN TANQKPKEQH KRNYVPCHIR QIINTWHKVG KNVYLPPREG DLTCNSTVTS LIANIDWIDG NQTNI TMSA EVAELYRLEL GDYKLVEITP IGLAPTSCKR YTTGGTSRRR RRR |
-Macromolecule #2: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 18.1446 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GVFVLGFLGF LATAGSAMGA ASLTLTAQSR TLLAGIVQQQ QQLLDVPKRQ QELLRLTVWG TKNLQTRVTA IEKYLKDQAQ LNAWGCAFR QVCCTTVPWP NASLIPKWNN ETWQEWERKV DFLEENITAL LEEAQIQQEK NMYELQKLNG SGHHHHHHHH |
-Macromolecule #3: K11 IgG heavy chain
| Macromolecule | Name: K11 IgG heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Macaca mulatta (Rhesus monkey) Macaca mulatta (Rhesus monkey) |
| Molecular weight | Theoretical: 53.29077 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DWTWRILFLV AAATGAHSQV QLQESGPGVV RPSQTLSLTC AVSGDTVSSC CFFWTWIRQP PGKGLEWIGN IYSDNDNTNY NPSLKTRIS ISKDMSKNQF SLKLNSLTAT DTAIYYCARE SPSRGNFCYA YLYGNCPLHF DLWGQGVLVT VSSASTKGPS V FPLAPSSR ...String: DWTWRILFLV AAATGAHSQV QLQESGPGVV RPSQTLSLTC AVSGDTVSSC CFFWTWIRQP PGKGLEWIGN IYSDNDNTNY NPSLKTRIS ISKDMSKNQF SLKLNSLTAT DTAIYYCARE SPSRGNFCYA YLYGNCPLHF DLWGQGVLVT VSSASTKGPS V FPLAPSSR STSESTAALG CLVKDYFPEP VTVSWNSGSL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYVCNVN HK PSNTKVD KRVEIKTCGG GSKPPTCPPC TSPELLGGPS VFLFPPKPKD TLMISRTPEV TCVVVDVSQE DPDVKFNWYV NGA EVHHAQ TKPRETQYNS TYRVVSVLTV THQDWLNGKE YTCKVSNKAL PAPIQKTISK DKGQPREPQV YTLPPSREEL TKNQ VSLTC LVKGFYPSDI VVEWESSGQP ENTYKTTPPV LDSDGSYFLY SKLTVDKSRW QQGNVFSCSV MHEALHNHYT QKSLS LSPG K |
-Macromolecule #4: K11 IgG light chain
| Macromolecule | Name: K11 IgG light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Macaca mulatta (Rhesus monkey) Macaca mulatta (Rhesus monkey) |
| Molecular weight | Theoretical: 25.29315 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ETDTLLLWVL LLWVPGSTGD ILLTQSPSSL SGSVGDRVTI TCRASQGINS YLNWYQQKPG KAPKLLIYFA NRLQSGVPSR FSGSGSGTE FTLTISSLQS EDGATYYCQQ YDTFPTFGPG TKLDIKRTVA APSVFIFPPS EDQVKSGTVS VVCLLNNFYP R EASVKWKV ...String: ETDTLLLWVL LLWVPGSTGD ILLTQSPSSL SGSVGDRVTI TCRASQGINS YLNWYQQKPG KAPKLLIYFA NRLQSGVPSR FSGSGSGTE FTLTISSLQS EDGATYYCQQ YDTFPTFGPG TKLDIKRTVA APSVFIFPPS EDQVKSGTVS VVCLLNNFYP R EASVKWKV DGALKTGNSQ ESVTEQDSKD NTYSLSSTLT LSSTEYQSHK VYACEVTHQG LSSPVTKSFN RGEC |
-Macromolecule #13: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 13 / Number of copies: 36 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7t4g: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)