+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BG505.MD39TS Env trimer in complex with Fab from antibody C05 | |||||||||
 Map data Map data | BG505.MD39TS Env trimer in complex with Fab from antibody C05 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |    Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
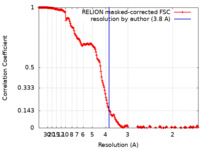
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.8 Å cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Moore A / Du J / Xu Z / Walker S / Kulp DW / Pallesen J | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Induction of tier-2 neutralizing antibodies in mice with a DNA-encoded HIV envelope native like trimer. Authors: Ziyang Xu / Susanne Walker / Megan C Wise / Neethu Chokkalingam / Mansi Purwar / Alan Moore / Edgar Tello-Ruiz / Yuanhan Wu / Sonali Majumdar / Kylie M Konrath / Abhijeet Kulkarni / Nicholas ...Authors: Ziyang Xu / Susanne Walker / Megan C Wise / Neethu Chokkalingam / Mansi Purwar / Alan Moore / Edgar Tello-Ruiz / Yuanhan Wu / Sonali Majumdar / Kylie M Konrath / Abhijeet Kulkarni / Nicholas J Tursi / Faraz I Zaidi / Emma L Reuschel / Ishaan Patel / April Obeirne / Jianqiu Du / Katherine Schultheis / Lauren Gites / Trevor Smith / Janess Mendoza / Kate E Broderick / Laurent Humeau / Jesper Pallesen / David B Weiner / Daniel W Kulp /  Abstract: HIV Envelope (Env) is the main vaccine target for induction of neutralizing antibodies. Stabilizing Env into native-like trimer (NLT) conformations is required for recombinant protein immunogens to ...HIV Envelope (Env) is the main vaccine target for induction of neutralizing antibodies. Stabilizing Env into native-like trimer (NLT) conformations is required for recombinant protein immunogens to induce autologous neutralizing antibodies(nAbs) against difficult to neutralize HIV strains (tier-2) in rabbits and non-human primates. Immunizations of mice with NLTs have generally failed to induce tier-2 nAbs. Here, we show that DNA-encoded NLTs fold properly in vivo and induce autologous tier-2 nAbs in mice. DNA-encoded NLTs also uniquely induce both CD4 + and CD8 + T-cell responses as compared to corresponding protein immunizations. Murine neutralizing antibodies are identified with an advanced sequencing technology. The structure of an Env-Ab (C05) complex, as determined by cryo-EM, identifies a previously undescribed neutralizing Env C3/V5 epitope. Beyond potential functional immunity gains, DNA vaccines permit in vivo folding of structured antigens and provide significant cost and speed advantages for enabling rapid evaluation of new HIV vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25376.map.gz emd_25376.map.gz | 479.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25376-v30.xml emd-25376-v30.xml emd-25376.xml emd-25376.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25376_fsc.xml emd_25376_fsc.xml | 18.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_25376.png emd_25376.png | 98.3 KB | ||
| Others |  emd_25376_additional_1.map.gz emd_25376_additional_1.map.gz emd_25376_half_map_1.map.gz emd_25376_half_map_1.map.gz emd_25376_half_map_2.map.gz emd_25376_half_map_2.map.gz | 410.6 MB 412.6 MB 410.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25376 http://ftp.pdbj.org/pub/emdb/structures/EMD-25376 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25376 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25376 | HTTPS FTP |
-Related structure data
| Related structure data |  7sq1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25376.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25376.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BG505.MD39TS Env trimer in complex with Fab from antibody C05 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_25376_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 1
| File | emd_25376_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_25376_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BG505.MD39TS Env trimer in complex with Fab from antibody C05
| Entire | Name: BG505.MD39TS Env trimer in complex with Fab from antibody C05 |
|---|---|
| Components |
|
-Supramolecule #1: BG505.MD39TS Env trimer in complex with Fab from antibody C05
| Supramolecule | Name: BG505.MD39TS Env trimer in complex with Fab from antibody C05 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Molecular weight | Theoretical: 410 KDa |
-Macromolecule #1: Transmembrane protein gp41
| Macromolecule | Name: Transmembrane protein gp41 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.134324 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVSLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEP QQHLLKDTHW GIKQLQARVL AVEHYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD |
-Macromolecule #2: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 52.917855 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ENLWVTVYYG VPVWKDAETT LFCASDAKAY ETEKHNVWAT HACVPTDPNP QEIHLENVTE EFNMWKNNMV EQMHEDIISL WDQSLKPCV KLTPLCVTLQ CTNVTNNITD DMRGELKNCS FNMTTELRDK KQKVYSLFYR LDVVQINENQ GNRSNNSNKE Y RLINCNTS ...String: ENLWVTVYYG VPVWKDAETT LFCASDAKAY ETEKHNVWAT HACVPTDPNP QEIHLENVTE EFNMWKNNMV EQMHEDIISL WDQSLKPCV KLTPLCVTLQ CTNVTNNITD DMRGELKNCS FNMTTELRDK KQKVYSLFYR LDVVQINENQ GNRSNNSNKE Y RLINCNTS AITQACPKVS FEPIPIHYCA PAGFAILKCK DKKFNGTGPC PSVSTVQCTH GIKPVVSTQL LLNGSLAEEE VI IRSENIT NNAKNILVQL NTPVQINCTR PNNNTVKSIR IGPGQAFYYT GDIIGDIRQA HCNVSKATWN ETLGKVVKQL RKH FGNNTI IRFAQSSGGD LEVTTHSFNC GGEFFYCNTS GLFNSTWISN TSVQGSNSTG SNDSITLPCR IKQIINMWQR IGQA MYAPP IQGVIRCVSN ITGLILTRDG GSTNSTTETF RPGGGDMRDN WRSELYKYKV VKIEPLGVAP TRCKRRV |
-Macromolecule #3: C05 Fab Light chain
| Macromolecule | Name: C05 Fab Light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 12.031465 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVLTQSPAS LAVSPGQRAT ISCRPSESVD NYGISFMNWF QQKPGQPPKL LIYAASNRGS GVPARFTGSG SGTDFSLNIH PMEEDDIAM YFCQQSKEVP YTFGGGTKLE IK |
-Macromolecule #4: C05 Fab heavy chain
| Macromolecule | Name: C05 Fab heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 13.727245 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVKLEESGGG LVQAGGSMKL SCVASGFSLS NYWMNWVRQS PEKGLEWVAE IRLKPQNYAT HYAESVKGRF SISRDDSRST VYLQMNNLR AEDTGIYYCT RPGYYGHYAM DYWGQGTSVT VSS |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 41 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 6000 pixel / Digitization - Dimensions - Height: 4000 pixel / Number grids imaged: 1 / Number real images: 5485 / Average exposure time: 2.59 sec. / Average electron dose: 60.6 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-7sq1: |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X