+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24513 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AAVrh.10-7x capsid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology | Phospholipase A2-like domain / Phospholipase A2-like domain / Parvovirus coat protein VP2 / Parvovirus coat protein VP1/VP2 / Parvovirus coat protein VP2 / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity /  Capsid protein VP1 Capsid protein VP1 Function and homology information Function and homology information | |||||||||
| Biological species |    Adeno-associated virus Adeno-associated virus | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.71 Å cryo EM / Resolution: 2.71 Å | |||||||||
 Authors Authors | Mietzsch M / McKenna R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2021 Journal: J Virol / Year: 2021Title: Structural Study of Aavrh.10 Receptor and Antibody Interactions. Authors: Mario Mietzsch / Jennifer C Yu / Jane Hsi / Paul Chipman / Felix Broecker / Zhang Fuming / Robert J Linhardt / Peter H Seeberger / Regine Heilbronn / Robert McKenna / Mavis Agbandje-McKenna /   Abstract: Recombinant adeno-associated virus (rAAV) vectors are one of the leading tools for the delivery of therapeutic genes in human gene therapy applications. For a successful transfer of their payload, ...Recombinant adeno-associated virus (rAAV) vectors are one of the leading tools for the delivery of therapeutic genes in human gene therapy applications. For a successful transfer of their payload, the AAV vectors have to circumvent potential preexisting neutralizing host antibodies and bind to the receptors of the target cells. Both of these aspects have not been structurally analyzed for AAVrh.10. Here, cryo-electron microscopy and three-dimensional image reconstruction were used to map the binding site of sulfated -acetyllactosamine (LacNAc; previously shown to bind AAVrh.10) and a series of four monoclonal antibodies (MAbs). LacNAc was found to bind to a pocket located on the side of the 3-fold capsid protrusion that is mostly conserved to AAV9 and equivalent to its galactose-binding site. As a result, AAVrh.10 was also shown to be able to bind to cell surface glycans with terminal galactose. For the antigenic characterization, it was observed that several anti-AAV8 MAbs cross-react with AAVrh.10. The binding sites of these antibodies were mapped to the 3-fold capsid protrusions. Based on these observations, the AAVrh.10 capsid surface was engineered to create variant capsids that escape these antibodies while maintaining infectivity. Gene therapy vectors based on adeno-associated virus rhesus isolate 10 (AAVrh.10) have been used in several clinical trials to treat monogenetic diseases. However, compared to other AAV serotypes little is known about receptor binding and antigenicity of the AAVrh.10 capsid. Particularly, preexisting neutralizing antibodies against capsids are an important challenge that can hamper treatment efficiency. This study addresses both topics and identifies critical regions of the AAVrh.10 capsid for receptor and antibody binding. The insights gained were utilized to generate AAVrh.10 variants capable of evading known neutralizing antibodies. The findings of this study could further aid the utilization of AAVrh.10 vectors in clinical trials and help the approval of the subsequent biologics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24513.map.gz emd_24513.map.gz | 226.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24513-v30.xml emd-24513-v30.xml emd-24513.xml emd-24513.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24513.png emd_24513.png | 73.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24513 http://ftp.pdbj.org/pub/emdb/structures/EMD-24513 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24513 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24513 | HTTPS FTP |
-Related structure data
| Related structure data |  7rl1MC  7s1wC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24513.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24513.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.049 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Adeno-associated virus
| Entire | Name:    Adeno-associated virus Adeno-associated virus |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus
| Supramolecule | Name: Adeno-associated virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 272636 / Sci species name: Adeno-associated virus / Sci species strain: rh.10-7x / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host system | Organism:   Homo sapiens (human) / Recombinant cell: HEK293 Homo sapiens (human) / Recombinant cell: HEK293 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Adeno-associated virus Adeno-associated virus |
| Molecular weight | Theoretical: 58.323359 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ADGVGSSSGN WHCDSTWLGD RVITTSTRTW ALPTYNNHLY KQISNGTSGG STNDNTYFGY STPWGYFDFN RFHCHFSPRD WQRLINNNW GFRPKRLNFK LFNIQVKEVT QNEGTKTIAN NLTSTIQVFT DSEYQLPYVL GSAHQGCLPP FPADVFMIPQ Y GYLTLNNG ...String: ADGVGSSSGN WHCDSTWLGD RVITTSTRTW ALPTYNNHLY KQISNGTSGG STNDNTYFGY STPWGYFDFN RFHCHFSPRD WQRLINNNW GFRPKRLNFK LFNIQVKEVT QNEGTKTIAN NLTSTIQVFT DSEYQLPYVL GSAHQGCLPP FPADVFMIPQ Y GYLTLNNG AQAVGRSSFY CLEYFPSQML RTGNNFEFSY QFEDVPFHSS YAHSQSLDRL MNPLIDQYLY YLSRTQTGGT AG TQQLLFS QAGPNNMSAQ AKNWLPGPCY RQQRVSTTLS QNNNSNFAWT GATKYHLNGR DSLVNPGVAM ATHKDDEERF FPS SGVLMF GKQGAGKDNV DYSAVMLTSE EEIKTTNPVA TEQYGVVADN LQQNSAQPIV GAVNSQGALP GMVWQNRDVY LQGP IWAKI PHTDGNFHPS PLMGGFGLKH PPPQILIKNT PVPADPPTTF SQAKLASFIT QYSTGQVSVE IEWELQKENS KRWNP EIQY TSNYYKSTNV DFAVNVDGTY SEPRPIGTRY LTRNL |
-Macromolecule #2: 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE
| Macromolecule | Name: 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE / type: ligand / ID: 2 / Number of copies: 60 / Formula: DC |
|---|---|
| Molecular weight | Theoretical: 307.197 Da |
| Chemical component information | 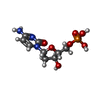 ChemComp-DC: |
-Macromolecule #3: 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE
| Macromolecule | Name: 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE / type: ligand / ID: 3 / Number of copies: 60 / Formula: DA |
|---|---|
| Molecular weight | Theoretical: 331.222 Da |
| Chemical component information | 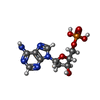 ChemComp-D5M: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm Bright-field microscopy / Cs: 2.7 mm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 75.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: cisTEM |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: RANDOM ASSIGNMENT |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.71 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM / Number images used: 17861 |
 Movie
Movie Controller
Controller






















