+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23015 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GluK2/K5 with L-Glu | |||||||||
 Map data Map data | GluK2/K5 with L-Glu (full-length map) | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of synaptic vesicle fusion to presynaptic active zone membrane / protein retention in ER lumen /  mossy fiber rosette / detection of cold stimulus involved in thermoception / Activation of Na-permeable kainate receptors / kainate selective glutamate receptor complex / Activation of Ca-permeable Kainate Receptor / negative regulation of synaptic transmission, glutamatergic / regulation of short-term neuronal synaptic plasticity / mossy fiber rosette / detection of cold stimulus involved in thermoception / Activation of Na-permeable kainate receptors / kainate selective glutamate receptor complex / Activation of Ca-permeable Kainate Receptor / negative regulation of synaptic transmission, glutamatergic / regulation of short-term neuronal synaptic plasticity /  inhibitory postsynaptic potential ...regulation of synaptic vesicle fusion to presynaptic active zone membrane / protein retention in ER lumen / inhibitory postsynaptic potential ...regulation of synaptic vesicle fusion to presynaptic active zone membrane / protein retention in ER lumen /  mossy fiber rosette / detection of cold stimulus involved in thermoception / Activation of Na-permeable kainate receptors / kainate selective glutamate receptor complex / Activation of Ca-permeable Kainate Receptor / negative regulation of synaptic transmission, glutamatergic / regulation of short-term neuronal synaptic plasticity / mossy fiber rosette / detection of cold stimulus involved in thermoception / Activation of Na-permeable kainate receptors / kainate selective glutamate receptor complex / Activation of Ca-permeable Kainate Receptor / negative regulation of synaptic transmission, glutamatergic / regulation of short-term neuronal synaptic plasticity /  inhibitory postsynaptic potential / inhibitory postsynaptic potential /  glutamate receptor activity / ubiquitin conjugating enzyme binding / receptor clustering / modulation of excitatory postsynaptic potential / neuronal action potential / regulation of JNK cascade / kainate selective glutamate receptor activity / glutamate receptor activity / ubiquitin conjugating enzyme binding / receptor clustering / modulation of excitatory postsynaptic potential / neuronal action potential / regulation of JNK cascade / kainate selective glutamate receptor activity /  ionotropic glutamate receptor complex / extracellularly glutamate-gated ion channel activity / behavioral fear response / positive regulation of synaptic transmission / glutamate-gated receptor activity / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / ionotropic glutamate receptor complex / extracellularly glutamate-gated ion channel activity / behavioral fear response / positive regulation of synaptic transmission / glutamate-gated receptor activity / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential /  excitatory postsynaptic potential / hippocampal mossy fiber to CA3 synapse / presynaptic modulation of chemical synaptic transmission / dendrite cytoplasm / excitatory postsynaptic potential / hippocampal mossy fiber to CA3 synapse / presynaptic modulation of chemical synaptic transmission / dendrite cytoplasm /  regulation of membrane potential / regulation of membrane potential /  bioluminescence / bioluminescence /  SNARE binding / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / generation of precursor metabolites and energy / SNARE binding / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / generation of precursor metabolites and energy /  synaptic transmission, glutamatergic / synaptic transmission, glutamatergic /  PDZ domain binding / establishment of localization in cell / cellular response to glucose stimulus / postsynaptic density membrane / regulation of long-term neuronal synaptic plasticity / modulation of chemical synaptic transmission / PDZ domain binding / establishment of localization in cell / cellular response to glucose stimulus / postsynaptic density membrane / regulation of long-term neuronal synaptic plasticity / modulation of chemical synaptic transmission /  terminal bouton / intracellular calcium ion homeostasis / terminal bouton / intracellular calcium ion homeostasis /  SH3 domain binding / positive regulation of neuron apoptotic process / SH3 domain binding / positive regulation of neuron apoptotic process /  presynaptic membrane / presynaptic membrane /  perikaryon / chemical synaptic transmission / perikaryon / chemical synaptic transmission /  postsynaptic membrane / postsynaptic membrane /  scaffold protein binding / neuron apoptotic process / negative regulation of neuron apoptotic process / scaffold protein binding / neuron apoptotic process / negative regulation of neuron apoptotic process /  postsynaptic density / postsynaptic density /  axon / axon /  dendrite / neuronal cell body / glutamatergic synapse / dendrite / neuronal cell body / glutamatergic synapse /  synapse / synapse /  ubiquitin protein ligase binding / ubiquitin protein ligase binding /  endoplasmic reticulum / endoplasmic reticulum /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Rattus norvegicus (Norway rat) / Rattus norvegicus (Norway rat) /   Aequorea victoria (jellyfish) Aequorea victoria (jellyfish) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 5.8 Å cryo EM / Resolution: 5.8 Å | |||||||||
 Authors Authors | Khanra N / Brown PMGE / Perozzo AM / Bowie D / Meyerson JR | |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor. Authors: Nandish Khanra / Patricia Mge Brown / Amanda M Perozzo / Derek Bowie / Joel R Meyerson /   Abstract: Kainate receptors (KARs) are L-glutamate-gated ion channels that regulate synaptic transmission and modulate neuronal circuits. KARs have strict assembly rules and primarily function as heteromeric ...Kainate receptors (KARs) are L-glutamate-gated ion channels that regulate synaptic transmission and modulate neuronal circuits. KARs have strict assembly rules and primarily function as heteromeric receptors in the brain. A longstanding question is how KAR heteromer subunits organize and coordinate together to fulfill their signature physiological roles. Here we report structures of the GluK2/GluK5 heteromer in apo, antagonist-bound, and desensitized states. The receptor assembles with two copies of each subunit, ligand binding domains arranged as two heterodimers and GluK5 subunits proximal to the channel. Strikingly, during desensitization, GluK2, but not GluK5, subunits undergo major structural rearrangements to facilitate channel closure. We show how the large conformational differences between antagonist-bound and desensitized states are mediated by the linkers connecting the pore helices to the ligand binding domains. This work presents the first KAR heteromer structure, reveals how its subunits are organized, and resolves how the heteromer can accommodate functionally distinct closed channel structures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23015.map.gz emd_23015.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23015-v30.xml emd-23015-v30.xml emd-23015.xml emd-23015.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
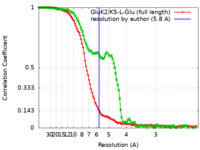
| FSC (resolution estimation) |  emd_23015_fsc.xml emd_23015_fsc.xml emd_23015_fsc_2.xml emd_23015_fsc_2.xml | 11.5 KB 11.2 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_23015.png emd_23015.png | 56.8 KB | ||
| Others |  emd_23015_additional_1.map.gz emd_23015_additional_1.map.gz emd_23015_additional_2.map.gz emd_23015_additional_2.map.gz | 96.9 MB 88.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23015 http://ftp.pdbj.org/pub/emdb/structures/EMD-23015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23015 | HTTPS FTP |
-Related structure data
| Related structure data |  7ks3MC  7ks0C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10660 (Title: GluK2/K5 with L-Glu / Data size: 3.4 TB / Data #1: GluK2/K5 with L-Glu [micrographs - multiframe] / Data #2: GluK2/K5 with L-Glu [micrographs - multiframe] EMPIAR-10660 (Title: GluK2/K5 with L-Glu / Data size: 3.4 TB / Data #1: GluK2/K5 with L-Glu [micrographs - multiframe] / Data #2: GluK2/K5 with L-Glu [micrographs - multiframe]Data #3: GluK2/K5 with L-Glu [picked particles - single frame - processed] Data #4: GluK2/K5 with L-Glu [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23015.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23015.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GluK2/K5 with L-Glu (full-length map) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.096 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: GluK2/K5 with L-Glu (ATD map)
| File | emd_23015_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GluK2/K5 with L-Glu (ATD map) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: GluK2/K5 with L-Glu (LBD-TMD map)
| File | emd_23015_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GluK2/K5 with L-Glu (LBD-TMD map) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GluK2/K5 with L-Glu
| Entire | Name: GluK2/K5 with L-Glu |
|---|---|
| Components |
|
-Supramolecule #1: GluK2/K5 with L-Glu
| Supramolecule | Name: GluK2/K5 with L-Glu / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK293S GnTI- Homo sapiens (human) / Recombinant cell: HEK293S GnTI- |
-Macromolecule #1: Glutamate receptor ionotropic, kainate 5,Green fluorescent protei...
| Macromolecule | Name: Glutamate receptor ionotropic, kainate 5,Green fluorescent protein chimera type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aequorea victoria (jellyfish) Aequorea victoria (jellyfish) |
| Molecular weight | Theoretical: 123.676812 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPAELLLLLI VAFANPSCQV LSSLRMAAIL DDQTVCGRGE RLALALAREQ INGIIEVPAK ARVEVDIFEL QRDSQYETTD TMCQILPKG VVSVLGPSSS PASASTVSHI CGEKEIPHIK VGPEETPRLQ YLRFASVSLY PSNEDVSLAV SRILKSFNYP S ASLICAKA ...String: MPAELLLLLI VAFANPSCQV LSSLRMAAIL DDQTVCGRGE RLALALAREQ INGIIEVPAK ARVEVDIFEL QRDSQYETTD TMCQILPKG VVSVLGPSSS PASASTVSHI CGEKEIPHIK VGPEETPRLQ YLRFASVSLY PSNEDVSLAV SRILKSFNYP S ASLICAKA ECLLRLEELV RGFLISKETL SVRMLDDSRD PTPLLKEIRD DKVSTIIIDA NASISHLVLR KASELGMTSA FY KYILTTM DFPILHLDGI VEDSSNILGF SMFNTSHPFY PEFVRSLNMS WRENCEASTY PGPALSAALM FDAVHVVVSA VRE LNRSQE IGVKPLACTS ANIWPHGTSL MNYLRMVEYD GLTGRVEFNS KGQRTNYTLR ILEKSRQGHR EIGVWYSNRT LAMN ATTLD INLSQTLANK TLVVTTILEN PYVMRRPNFQ ALSGNERFEG FCVDMLRELA ELLRFRYRLR LVEDGLYGAP EPNGS WTGM VGELINRKAD LAVAAFTITA EREKVIDFSK PFMTLGISIL YRVHMGRKPG YFSFLDPFSP AVWLFMLLAY LAVSVV LFL AARLSPYEWY NPHPSLRARP HILENQYTLG NSLWFPVGGF MQQGSEIMPR ALSTRIVSGV WWAFTLIIIS SYTANLA AF LTVQRMEVPV ESADDLADQT NIEYGTIHAG STMTFFQNSR YQTYQRMWNY MQSKQPSVFV KSTEEGIARV LNSRYAFL L ESTMNEYHRR LNCNLTQIGG LLDTKGYGIG MPLGSPFRDE ITLAILQLQE NNRLEILKRK WWEGGRCPKE EDHRAKGLG MENIGGIFVV LIAGLIIAVF VAVMEFIYKS RAEAKRMKGL VPRGSAAAAM VSKGEELFTG VVPILVELDG DVNGHKFSVS GEGEGDATY GKLTLKFICT TGKLPVPWPT LVTTLTYGVQ CFSRYPDHMK QHDFFKSAMP EGYVQERTIF FKDDGNYKTR A EVKFEGDT LVNRIELKGI DFKEDGNILG HKLEYNYNSH NVYIMADKQK NGIKVNFKIR HNIEDGSVQL ADHYQQNTPI GD GPVLLPD NHYLSTQSKL SKDPNEKRDH MVLLEFVTAA GITLGMDELY KSGLRTETSQ VAPA |
-Macromolecule #2: Glutamate receptor ionotropic, kainate 2
| Macromolecule | Name: Glutamate receptor ionotropic, kainate 2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) |
| Molecular weight | Theoretical: 105.981617 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKIISPVLSN LVFSRSIKVL LCLLWIGYSQ GTTHVLRFGG IFEYVESGPM GAEELAFRFA VNTINRNRTL LPNTTLTYDT QKINLYDSF EASKKACDQL SLGVAAIFGP SHSSSANAVQ SICNALGVPH IQTRWKHQVS DNKDSFYVSL YPDFSSLSRA I LDLVQFFK ...String: MKIISPVLSN LVFSRSIKVL LCLLWIGYSQ GTTHVLRFGG IFEYVESGPM GAEELAFRFA VNTINRNRTL LPNTTLTYDT QKINLYDSF EASKKACDQL SLGVAAIFGP SHSSSANAVQ SICNALGVPH IQTRWKHQVS DNKDSFYVSL YPDFSSLSRA I LDLVQFFK WKTVTVVYDD STGLIRLQEL IKAPSRYNLR LKIRQLPADT KDAKPLLKEM KRGKEFHVIF DCSHEMAAGI LK QALAMGM MTEYYHYIFT TLDLFALDVE PYRYSGVNMT GFRILNTENT QVSSIIEKWS MERLQAPPKP DSGLLDGFMT TDA ALMYDA VHVVSVAVQQ FPQMTVSSLQ CNRHKPWRFG TRFMSLIKEA HWEGLTGRIT FNKTNGLRTD FDLDVISLKE EGLE KIGTW DPASGLNMTE SQKGKPANIT DSLSNRSLIV TTILEEPYVL FKKSDKPLYG NDRFEGYCID LLRELSTILG FTYEI RLVE DGKYGAQDDV NGQWNGMVRE LIDHKADLAV APLAITYVRE KVIDFSKPFM TLGISILYRK PNGTNPGVFS FLNPLS PDI WMYVLLAYLG VSVVLFVIAR FSPYEWYNPH PSNPDSDVVE NNFTLLNSFW FGVGALMQQG SELMPKALST RIVGGIW WF FTLIIISSYT ANLAAFLTVE RMESPIDSAD DLAKQTKIEY GAVEDGATMT FFKKSKISTY DKMWAFMSSR RQSVLVKS N EEGIQRVLTS DYAFLMESTT IEFVTQRNCN LTQIGGLIDS KGYGVGTPMG SPYRDKITIA ILQLQEEGKL HMMKEKWWR GNGCPEEESK EASALGVQNI GGIFIVLAAG LVLSVFVAVG EFLYKSKKNA QLEKRSFCSA MVEELRMSLK CQRRLKHKPQ APVIVKTEE VINMHTFNDR RLPGKETMAS GLRSAWSHPQ FEKGGGSGGG SGGGSWSHPQ FEK |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.7 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | 1 mM L-Glu |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 51.5 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z
Z Y
Y X
X