+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22990 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the H-lobe of yeast CKM | |||||||||
 Map data Map data | Structure of the H-lobe of yeast CKM | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mediator /  Transcription / Transcription /  Cdk8 / Cdk8 /  Med13 / Med13 /  Med12 / CycC / CDK / Med12 / CycC / CDK /  Argonaute / Argonaute /  RNA Polymerase II / RNA Polymerase II /  PIWI PIWI | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of transcription by galactose / CKM complex /  mediator complex / mediator complex /  transcription coactivator activity / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II transcription coactivator activity / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase IISimilarity search - Function | |||||||||
| Biological species |   Saccharomyces cerevisiae S288C (yeast) / Saccharomyces cerevisiae S288C (yeast) /   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) | |||||||||
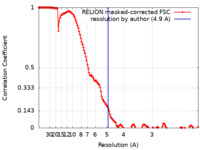
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.9 Å cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Li YC / Chao TC | |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module. Authors: Yi-Chuan Li / Ti-Chun Chao / Hee Jong Kim / Timothy Cholko / Shin-Fu Chen / Guojie Li / Laura Snyder / Kotaro Nakanishi / Chia-En Chang / Kenji Murakami / Benjamin A Garcia / Thomas G Boyer / Kuang-Lei Tsai /  Abstract: The Cdk8 kinase module (CKM) in Mediator, comprising Med13, Med12, CycC, and Cdk8, regulates RNA polymerase II transcription through kinase-dependent and -independent functions. Numerous pathogenic ...The Cdk8 kinase module (CKM) in Mediator, comprising Med13, Med12, CycC, and Cdk8, regulates RNA polymerase II transcription through kinase-dependent and -independent functions. Numerous pathogenic mutations causative for neurodevelopmental disorders and cancer congregate in CKM subunits. However, the structure of the intact CKM and the mechanism by which Cdk8 is non-canonically activated and functionally affected by oncogenic CKM alterations are poorly understood. Here, we report a cryo-electron microscopy structure of CKM that redefines prior CKM structural models and explains the mechanism of Med12-dependent Cdk8 activation. Med12 interacts extensively with CycC and activates Cdk8 by stabilizing its activation (T-)loop through conserved Med12 residues recurrently mutated in human tumors. Unexpectedly, Med13 has a characteristic Argonaute-like bi-lobal architecture. These findings not only provide a structural basis for understanding CKM function and pathological dysfunction, but also further impute a previously unknown regulatory mechanism of Mediator in transcriptional modulation through its Med13 Argonaute-like features. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22990.map.gz emd_22990.map.gz | 9.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22990-v30.xml emd-22990-v30.xml emd-22990.xml emd-22990.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22990_fsc.xml emd_22990_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_22990.png emd_22990.png | 46.5 KB | ||
| Filedesc metadata |  emd-22990.cif.gz emd-22990.cif.gz | 6.2 KB | ||
| Others |  emd_22990_half_map_1.map.gz emd_22990_half_map_1.map.gz emd_22990_half_map_2.map.gz emd_22990_half_map_2.map.gz | 171 MB 171 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22990 http://ftp.pdbj.org/pub/emdb/structures/EMD-22990 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22990 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22990 | HTTPS FTP |
-Related structure data
| Related structure data |  7kpwMC  7kpvC  7kpxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22990.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22990.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the H-lobe of yeast CKM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half-volume 1
| File | emd_22990_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-volume 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half-volume 2
| File | emd_22990_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-volume 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : yeast CDK8 complex
| Entire | Name: yeast CDK8 complex Cyclin-dependent kinase 8 Cyclin-dependent kinase 8 |
|---|---|
| Components |
|
-Supramolecule #1: yeast CDK8 complex
| Supramolecule | Name: yeast CDK8 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae S288C (yeast) Saccharomyces cerevisiae S288C (yeast) |
| Molecular weight | Theoretical: 430 KDa |
-Macromolecule #1: Mediator of RNA polymerase II transcription subunit 12
| Macromolecule | Name: Mediator of RNA polymerase II transcription subunit 12 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast)Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 167.049812 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae S288C (yeast) Saccharomyces cerevisiae S288C (yeast) |
| Sequence | String: MNNGSGRYLL TPPDDLHPYV PSSKPQEQVY PDFKPWEHTA AEDQILANFV AKGFYHTPMV NFESISARSS VHESLVTQSN ILSQQFDKI IKIREDHINK IPSNSTTTLH GPGFQLPNRI TLTDHRKETW LHELSSSHTS LVKIGKFIPH GLKRRQVIEQ C YLKFIPLK ...String: MNNGSGRYLL TPPDDLHPYV PSSKPQEQVY PDFKPWEHTA AEDQILANFV AKGFYHTPMV NFESISARSS VHESLVTQSN ILSQQFDKI IKIREDHINK IPSNSTTTLH GPGFQLPNRI TLTDHRKETW LHELSSSHTS LVKIGKFIPH GLKRRQVIEQ C YLKFIPLK RAIWLIKCCY FIEWKSNHKK KRSNAAGADD AISMHLLKDW TDTFVYILEK LIFDMTNHYN DSQQLRTWKR QI SYFLKLL GNCYSLRLIN KEIFHHWLVE FINKMENFEF LPLSLHILMI FWNDICQIDT NAPVAATITS SQKEPFFLVT KIT DMLLHK YYIVSSSKSM INDENYIIND IKKNNKIKLN ILKILSSLIL KIFQEQSLEV FIFPTSNWEI YKPLLFEIVS NADT NQNSD MKKKLELISY RNESLKNNSS IRNVIMSASN ANDFQLTIVT CKQFPKLSCI QLNCIDTQFT KLLDDNPTEF DWPTY VDQN PLTMHKIIQL ILWSIHPSRQ FDHYESNQLV AKLLLLRINS TDEDLHEFQI EDAIWSLVFQ LAKNFSAQKR VVSYMM PSL YRLLNILITY GIIKVPTYIR KLISSGLLYL QDSNDKFVHV QLLINLKISP LMKSQYNMVL RNVMEYDVKF YEIFNFD QL VEITEQIKMR ILSNDITNLQ LSKTPLSIKI MVAEWYLSHL CSGILSSVNR TVLLKIFKIF CIDLEVFHHF FKWIEFIV Y HQLLSDIESL EALMDILLCY QKLFSQFIND HILFTKTFIF IYKKVLKEKD VPAYNVTSFM PFWKFFMKNF PFVLKVDND LRIELQSVYN DEKLKTEKLK NDKSEVLKVY SMINNSNQAV GQTWNFPEVF QVNIRFLLHN SEIIDTNTSK QFQKARNNVM LLIATNLKE YNKFMSIFLK RKDFTNKNLI QLISLKLLTF EVTQNVLGLE YIIRLLPINL ENNDGSYGLF LKYHKEQFIK S NFEKILLT CYELEKKYHG NECEINYYEI LLKILITYGS SPKLLATSTK IIMLLLNDSV ENSSNILEDI LYYSTCPSET DL NDIPLGS GQPDNDTVVT NDDKSDDDDH TVDEIDHVEY YVMMDFANLW VFQAFTCFCI KKIMENNEPA MAMEDLKNFI FQI IEITNS NDLCSQIFDQ LKDMQTIEMI TQIVEKDFCT SCLQNNNQKI DDNYIVVVIE IITSLSMRFQ RETSGMIVIS MENY HLLIK IIRQLSELNE GNLSKREIQI DAVLKIFSFH QDSIFQRIIA DLSADKPTSP FIDSICKLFD KISFNLRLKL FLYEI LSSL KSFAIYSSTI DAPAFHTSGK VELPKKLLNL PPFQVSSFVK ETKLHSGDYG EEEDADQEES FSLNLGIGIV EIAHEN EQK WLIYDKKDHK YVCTFSMEPY HFISNYNTKY TDDMATGSND TTAFNDSCVN LSLFDARFER KNPH UniProtKB: Mediator of RNA polymerase II transcription subunit 12 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 25 mM Hepes pH 7.4, 200 mM NaCl, and 0.005% NP-40 |
| Grid | Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm Bright-field microscopy / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number real images: 15075 / Average electron dose: 65.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z
Z Y
Y X
X