[English] 日本語
 Yorodumi
Yorodumi- EMDB-19039: Map of YPEL5-bound WDR26 dimer obtained by focused refinement of ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Map of YPEL5-bound WDR26 dimer obtained by focused refinement of the WDR26-CTLH subcomplex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GID / CTLH / WDR26 / YPEL5 /  ubiquitin / ubiquitin /  E3 ligase / E3 ligase /  supramolecular assembly / supramolecular assembly /  metabolism / metabolism /  gluconeogenesis / gluconeogenesis /  cryoEM / cryoEM /  LIGASE LIGASE | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
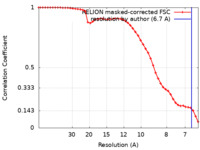
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 6.7 Å cryo EM / Resolution: 6.7 Å | |||||||||
 Authors Authors | Chrustowicz J / Schulman BA | |||||||||
| Funding support | European Union,  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: FEBS Lett / Year: 2024 Journal: FEBS Lett / Year: 2024Title: Skraban-Deardorff intellectual disability syndrome-associated mutations in WDR26 impair CTLH E3 complex assembly. Authors: Annette Gross / Judith Müller / Jakub Chrustowicz / Alexander Strasser / Karthik V Gottemukkala / Dawafuti Sherpa / Brenda A Schulman / Peter J Murray / Arno F Alpi /  Abstract: Patients with Skraban-Deardorff syndrome (SKDEAS), a neurodevelopmental syndrome associated with a spectrum of developmental and intellectual delays and disabilities, harbor diverse mutations in ...Patients with Skraban-Deardorff syndrome (SKDEAS), a neurodevelopmental syndrome associated with a spectrum of developmental and intellectual delays and disabilities, harbor diverse mutations in WDR26, encoding a subunit of the multiprotein CTLH E3 ubiquitin ligase complex. Structural studies revealed that homodimers of WDR26 bridge two core-CTLH E3 complexes to generate giant, hollow oval-shaped supramolecular CTLH E3 assemblies. Additionally, WDR26 mediates CTLH E3 complex binding to subunit YPEL5 and functions as substrate receptor for the transcriptional repressor HBP1. Here, we mapped SKDEAS-associated mutations on a WDR26 structural model and tested their functionality in complementation studies using genetically engineered human cells lacking CTLH E3 supramolecular assemblies. Despite the diversity of mutations, 15 of 16 tested mutants impaired at least one CTLH E3 complex function contributing to complex assembly and interactions, thus providing first mechanistic insights into SKDEAS pathology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19039.map.gz emd_19039.map.gz | 671 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19039-v30.xml emd-19039-v30.xml emd-19039.xml emd-19039.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19039_fsc.xml emd_19039_fsc.xml | 4.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_19039.png emd_19039.png | 157.3 KB | ||
| Masks |  emd_19039_msk_1.map emd_19039_msk_1.map | 8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19039.cif.gz emd-19039.cif.gz | 4.1 KB | ||
| Others |  emd_19039_half_map_1.map.gz emd_19039_half_map_1.map.gz emd_19039_half_map_2.map.gz emd_19039_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19039 http://ftp.pdbj.org/pub/emdb/structures/EMD-19039 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19039 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19039 | HTTPS FTP |
-Related structure data
| Related structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19039.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19039.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.214 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19039_msk_1.map emd_19039_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19039_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19039_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : WDR26-CTLH E3 subcomplex comprising RANBP9, TWA1, ARMC8, GID4, WD...
| Entire | Name: WDR26-CTLH E3 subcomplex comprising RANBP9, TWA1, ARMC8, GID4, WDR26 and YPEL5 |
|---|---|
| Components |
|
-Supramolecule #1: WDR26-CTLH E3 subcomplex comprising RANBP9, TWA1, ARMC8, GID4, WD...
| Supramolecule | Name: WDR26-CTLH E3 subcomplex comprising RANBP9, TWA1, ARMC8, GID4, WDR26 and YPEL5 type: complex / ID: 1 / Parent: 0 Details: Map obtained by focused refinement of the previously published map (EMD-12545) over the YPEL5-bound WDR26 dimer |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 300 KDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 75.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)