[English] 日本語
 Yorodumi
Yorodumi- EMDB-18886: Focused map on the Roc-COR domains of the Roco protein from C. te... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focused map on the Roc-COR domains of the Roco protein from C. tepidum in the GTP state bound to the activating Nanobody NbRoco1 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  GTPase / GTPase /  Nanobody / Nanobody /  Parkinson's Disease / Allostery activator / Parkinson's Disease / Allostery activator /  HYDROLASE HYDROLASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |   Chlorobaculum tepidum (bacteria) / Chlorobaculum tepidum (bacteria) /   Lama glama (llama) Lama glama (llama) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.88 Å cryo EM / Resolution: 3.88 Å | ||||||||||||
 Authors Authors | Galicia C / Versees W | ||||||||||||
| Funding support |  Belgium, 3 items Belgium, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structural insights into the GTP-driven monomerization and activation of a bacterial LRRK2 homolog using allosteric nanobodies. Authors: Christian Galicia / Giambattista Guaitoli / Marcus Fislage / Christian Johannes Gloeckner / Wim Versées /   Abstract: Roco proteins entered the limelight after mutations in human LRRK2 were identified as a major cause of familial Parkinson's disease. LRRK2 is a large and complex protein combining a GTPase and ...Roco proteins entered the limelight after mutations in human LRRK2 were identified as a major cause of familial Parkinson's disease. LRRK2 is a large and complex protein combining a GTPase and protein kinase activity, and disease mutations increase the kinase activity, while presumably decreasing the GTPase activity. Although a cross-communication between both catalytic activities has been suggested, the underlying mechanisms and the regulatory role of the GTPase domain remain unknown. Several structures of LRRK2 have been reported, but structures of Roco proteins in their activated GTP-bound state are lacking. Here, we use single-particle cryo-electron microscopy to solve the structure of a bacterial Roco protein (CtRoco) in its GTP-bound state, aided by two conformation-specific nanobodies: Nb and Nb. This structure presents CtRoco in an active monomeric state, featuring a very large GTP-induced conformational change using the LRR-Roc linker as a hinge. Furthermore, this structure shows how Nb and Nb collaborate to activate CtRoco in an allosteric way. Altogether, our data provide important new insights into the activation mechanism of Roco proteins, with relevance to LRRK2 regulation, and suggest new routes for the allosteric modulation of their GTPase activity. #1:  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Structural insights in the GTP-driven monomerization and activation of a bacterial LRRK2 homologue using allosteric nanobodies Authors: Galicia C / Guaitoli G / Fislage M / Gloeckner CJ / Versees W | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18886.map.gz emd_18886.map.gz | 421.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18886-v30.xml emd-18886-v30.xml emd-18886.xml emd-18886.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18886_fsc.xml emd_18886_fsc.xml | 5.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18886.png emd_18886.png | 94.2 KB | ||
| Masks |  emd_18886_msk_1.map emd_18886_msk_1.map | 18.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18886.cif.gz emd-18886.cif.gz | 6.5 KB | ||
| Others |  emd_18886_half_map_1.map.gz emd_18886_half_map_1.map.gz emd_18886_half_map_2.map.gz emd_18886_half_map_2.map.gz | 16.8 MB 16.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18886 http://ftp.pdbj.org/pub/emdb/structures/EMD-18886 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18886 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18886 | HTTPS FTP |
-Related structure data
| Related structure data |  8r4dMC  8r4bC  8r4cC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18886.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18886.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.4381 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18886_msk_1.map emd_18886_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
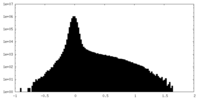
| Density Histograms |
-Half map: #2
| File | emd_18886_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
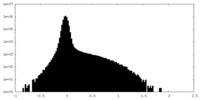
| Density Histograms |
-Half map: #1
| File | emd_18886_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CtRoco in the active GTP state bound to the two activating Nanobo...
| Entire | Name: CtRoco in the active GTP state bound to the two activating Nanobodies NbRoco1 and NbRoco2 |
|---|---|
| Components |
|
-Supramolecule #1: CtRoco in the active GTP state bound to the two activating Nanobo...
| Supramolecule | Name: CtRoco in the active GTP state bound to the two activating Nanobodies NbRoco1 and NbRoco2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Chlorobaculum tepidum (bacteria) Chlorobaculum tepidum (bacteria) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Rab family protein
| Macromolecule | Name: Rab family protein / type: protein_or_peptide / ID: 1 / Details: CtRoco bound to GTPgammaS / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Chlorobaculum tepidum (bacteria) Chlorobaculum tepidum (bacteria) |
| Molecular weight | Theoretical: 124.313453 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: GAMGSMSDLD VIRQIEQELG MQLEPVDKLK WYSKGYKLDK DQRVTAIGLY DCGSDTLDRI IQPLESLKSL SELSLSSNQI TDISPLASL NSLSMLWLDR NQITDIAPLA SLNSLSMLWL FGNKISDIAP LESLKSLTEL QLSSNQITDI APLASLKSLT E LSLSGNNI ...String: GAMGSMSDLD VIRQIEQELG MQLEPVDKLK WYSKGYKLDK DQRVTAIGLY DCGSDTLDRI IQPLESLKSL SELSLSSNQI TDISPLASL NSLSMLWLDR NQITDIAPLA SLNSLSMLWL FGNKISDIAP LESLKSLTEL QLSSNQITDI APLASLKSLT E LSLSGNNI SDIAPLESLK SLTELSLSSN QITDIAPLAS LKSLTELSLS SNQISDIAPL ESLKSLTELQ LSRNQISDIA PL ESLKSLT ELQLSSNQIT DIAPLASLKS LTELQLSRNQ ISDIAPLESL NSLSKLWLNG NQITDIAPLA SLNSLTELEL SSN QITDIA PLASLKSLST LWLSSNQISD IAPLASLESL SELSLSSNQI SDISPLASLN SLTGFDVRRN PIKRLPETIT GFDM EILWN DFSSSGFITF FDNPLESPPP EIVKQGKEAV RQYFQSIEEA RSKGEALVHL QEIKVHLIGD GMAGKTSLLK QLIGE TFDP KESQTHGLNV VTKQAPNIKG LENDDELKEC LFHFWDFGGQ EIMHASHQFF MTRSSVYMLL LDSRTDSNKH YWLRHI EKY GGKSPVIVVM NKIDENPSYN IEQKKINERF PAIENRFHRI SCKNGDGVES IAKSLKSAVL HPDSIYGTPL APSWIKV KE KLVEATTAQR YLNRTEVEKI CNDSGITDPG ERKTLLGYLN NLGIVLYFEA LDLSEIYVLD PHWVTIGVYR IINSSKTK N GHLNTSALGY ILNEEQIRCD EYDPAKNNKF TYTLLEQRYL LDIMKQFELC YDEGKGLFII PSNLPTQIDN EPEITEGEP LRFIMKYDYL PSTIIPRLMI AMQHQILDRM QWRYGMVLKS QDHEGALAKV VAETKDSTIT IAIQGEPRCK REYLSIIWYE IKKINANFT NLDVKEFIPL PGHPDELVEY KELLGLEKMG RDEYVSGKLE KVFSVSKMLD SVISKEERNK ERLMGDINIK L ENIGNPTI PIHQQVEVNV SQETVQHVEN LQGFFENLKA DILREAELEI DDPKERKRLA NELELAENAI TKMDAAVKSG KN KLKPDVK DRLGEFIDNL ANENSRLRKG IALVMNGAEK VQKLARYYNN VAPFFDLPSV PPVLLGKEKT UniProtKB: Rab family protein |
-Macromolecule #2: NbRoco1
| Macromolecule | Name: NbRoco1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Lama glama (llama) Lama glama (llama) |
| Molecular weight | Theoretical: 15.206718 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: QVQLQESGGG LVQAGGSLRL SCANSGLTFS TYTMGWFRQA PGKEREFVAA IRWSGTSTYY QDHADSVKGR FTISRDNAKN TVYLQMNSL KPEDTAVYYC AASRLRAGVK APSEYDYWGQ GTQVTVSSHH HHHHEPEA |
-Macromolecule #3: 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE
| Macromolecule | Name: 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: GSP |
|---|---|
| Molecular weight | Theoretical: 539.246 Da |
| Chemical component information |  ChemComp-GSP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.08 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.55 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 60000 Bright-field microscopy / Cs: 2.55 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 60000 |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 63.0 e/Å2 |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Protocol: FLEXIBLE FIT | ||||||
| Output model |  PDB-8r4d: |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X