[English] 日本語
 Yorodumi
Yorodumi- EMDB-18879: Roco protein from C. tepidum in the GTP state bound to an activat... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Roco protein from C. tepidum in the GTP state bound to an activating Nanobody | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  GTPase / GTPase /  Nanobody / Nanobody /  Parkinson's Disease / Allostery activator / Parkinson's Disease / Allostery activator /  HYDROLASE HYDROLASE | ||||||||||||
| Biological species |   Chlorobaculum tepidum (bacteria) / Chlorobaculum tepidum (bacteria) /   Lama glama (llama) Lama glama (llama) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 8.3 Å cryo EM / Resolution: 8.3 Å | ||||||||||||
 Authors Authors | Galicia C / Fislage M / Versees W | ||||||||||||
| Funding support |  Belgium, 3 items Belgium, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structural insights into the GTP-driven monomerization and activation of a bacterial LRRK2 homolog using allosteric nanobodies. Authors: Christian Galicia / Giambattista Guaitoli / Marcus Fislage / Christian Johannes Gloeckner / Wim Versées /   Abstract: Roco proteins entered the limelight after mutations in human LRRK2 were identified as a major cause of familial Parkinson's disease. LRRK2 is a large and complex protein combining a GTPase and ...Roco proteins entered the limelight after mutations in human LRRK2 were identified as a major cause of familial Parkinson's disease. LRRK2 is a large and complex protein combining a GTPase and protein kinase activity, and disease mutations increase the kinase activity, while presumably decreasing the GTPase activity. Although a cross-communication between both catalytic activities has been suggested, the underlying mechanisms and the regulatory role of the GTPase domain remain unknown. Several structures of LRRK2 have been reported, but structures of Roco proteins in their activated GTP-bound state are lacking. Here, we use single-particle cryo-electron microscopy to solve the structure of a bacterial Roco protein (CtRoco) in its GTP-bound state, aided by two conformation-specific nanobodies: Nb and Nb. This structure presents CtRoco in an active monomeric state, featuring a very large GTP-induced conformational change using the LRR-Roc linker as a hinge. Furthermore, this structure shows how Nb and Nb collaborate to activate CtRoco in an allosteric way. Altogether, our data provide important new insights into the activation mechanism of Roco proteins, with relevance to LRRK2 regulation, and suggest new routes for the allosteric modulation of their GTPase activity. #1:  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Structural insights in the GTP-driven monomerization and activation of a bacterial LRRK2 homologue using allosteric nanobodies Authors: Galicia C / Guaitoli G / Fislage M / Gloeckner CJ / Versees W | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18879.map.gz emd_18879.map.gz | 31.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18879-v30.xml emd-18879-v30.xml emd-18879.xml emd-18879.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18879_fsc.xml emd_18879_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18879.png emd_18879.png | 50.6 KB | ||
| Masks |  emd_18879_msk_1.map emd_18879_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18879.cif.gz emd-18879.cif.gz | 5.5 KB | ||
| Others |  emd_18879_half_map_1.map.gz emd_18879_half_map_1.map.gz emd_18879_half_map_2.map.gz emd_18879_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18879 http://ftp.pdbj.org/pub/emdb/structures/EMD-18879 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18879 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18879 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18879.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18879.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2415 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18879_msk_1.map emd_18879_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18879_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18879_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CtRoco in the active GTP state bound to the activating Nanobody N...
| Entire | Name: CtRoco in the active GTP state bound to the activating Nanobody NbRoco1 |
|---|---|
| Components |
|
-Supramolecule #1: CtRoco in the active GTP state bound to the activating Nanobody N...
| Supramolecule | Name: CtRoco in the active GTP state bound to the activating Nanobody NbRoco1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Chlorobaculum tepidum (bacteria) Chlorobaculum tepidum (bacteria) |
| Molecular weight | Theoretical: 140 KDa |
-Macromolecule #1: CtRoco
| Macromolecule | Name: CtRoco / type: protein_or_peptide / ID: 1 / Details: CtRoco bound to GTPgammaS / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Chlorobaculum tepidum (bacteria) Chlorobaculum tepidum (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: GAMGSMSDLD VIRQIEQELG MQLEPVDKLK WYSKGYKLDK DQRVTAIGLY DCGSDTLDRI IQP LESLKS LSELSLSSNQ ITDISPLASL NSLSMLWLDR NQITDIAPLA SLNSLSMLWL FGNKISDIAP L ESLKSLTE LQLSSNQITD IAPLASLKSL TELSLSGNNI ...String: GAMGSMSDLD VIRQIEQELG MQLEPVDKLK WYSKGYKLDK DQRVTAIGLY DCGSDTLDRI IQP LESLKS LSELSLSSNQ ITDISPLASL NSLSMLWLDR NQITDIAPLA SLNSLSMLWL FGNKISDIAP L ESLKSLTE LQLSSNQITD IAPLASLKSL TELSLSGNNI SDIAPLESLK SLTELSLSSN QITDIAPLAS L KSLTELSL SSNQISDIAP LESLKSLTEL QLSRNQISDI APLESLKSLT ELQLSSNQIT DIAPLASLKS L TELQLSRN QISDIAPLES LNSLSKLWLN GNQITDIAPL ASLNSLTELE LSSNQITDIA PLASLKSLS TLWLSSNQIS DIAPLASLES LSELSLSSNQ ISDISPLASL NSLTGFDVRR NPIKRLPETI TGFDMEIL W NDFSSSGFIT FFDNPLESPP PEIVKQGKEA VRQYFQSIEE ARSKGEALVH LQEIKVHLIG DGMA GKTSL LKQLIGETFD PKESQTHGLN VVTKQAPNIK GLENDDELKE CLFHFWDFGG QEIMHASH Q FFMTRSSVYM LLLDSRTDSN KHYWLRHIEK YGGKSPVIVV MNKIDENPSY NIEQKKINER FPA IENRFH RISCKNGDGV ESIAKSLKSA VLHPDSIYGT PLAPSWIKVK EKLVEATTAQ RYLNRTEVE KICNDSGITD PGERKTLLGY LNNLGIVLYF EALDLSEIYV LDPHWVTIGV YRIINSSKTK NGHLN TSAL GYILNEEQIR CDEYDPAKNN KFTYTLLEQR YLLDIMKQFE LCYDEGKGLF IIPSNLPTQI D NEPEITEG EPLRFIMKYD YLPSTIIPRL MIAMQHQILD RMQWRYGMVL KSQDHEGALA KVVAE TKDS TITIAIQGEP RCKREYLSII WYEIKKINAN FTNLDVKEFI PLPGHPDELV EYKELLGLEK MG RDEYVSG KLEKVFSVSK MLDSVISKEE RNKERLMGDI NIKLENIGNP TIPIHQQVEV NVSQETV QH VENLQGFFEN LKADILREAE LEIDDPKERK RLANELELAE NAITKMDAAV KSGKNKLKPD V KDRLGEFI DNLANENSRL RKGIALVMNG AEKVQKLARY YNNVAPFFDL PSVPPVLLGK EKT |
-Macromolecule #2: NbRoco1
| Macromolecule | Name: NbRoco1 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Lama glama (llama) Lama glama (llama) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: QVQLQESGGG LVQAGGSLRL SCANSGLTFS TYTMGWFRQA PGKEREFVAA IRWSGTSTYY QDHADSVKGR FTISRDNAKN TVYLQMNSLK PEDTAVYYCA ASRLRAGVKA PSEYDYWGQG TQVTVSSHHH HHHEPEA |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.08 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.55 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 60000 Bright-field microscopy / Cs: 2.55 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 60000 |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 63.0 e/Å2 |
 Movie
Movie Controller
Controller






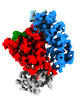



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)