[English] 日本語
 Yorodumi
Yorodumi- EMDB-18806: Structural insights into the activation mechanism of antimicrobia... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural insights into the activation mechanism of antimicrobial GBP1: Membrane-bound GBP1 oligomer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Oligomer / Oligomer /  GTPase / Interferon-induced / GTPase / Interferon-induced /  ANTIMICROBIAL PROTEIN ANTIMICROBIAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 26.8 Å cryo EM / Resolution: 26.8 Å | |||||||||
 Authors Authors | Weismehl M / Chu X / Kutsch M / Lauterjung P / Herrmann C / Kudryashev M / Daumke O | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2024 Journal: EMBO J / Year: 2024Title: Structural insights into the activation mechanism of antimicrobial GBP1. Authors: Marius Weismehl / Xiaofeng Chu / Miriam Kutsch / Paul Lauterjung / Christian Herrmann / Misha Kudryashev / Oliver Daumke /   Abstract: The dynamin-related human guanylate-binding protein 1 (GBP1) mediates host defenses against microbial pathogens. Upon GTP binding and hydrolysis, auto-inhibited GBP1 monomers dimerize and assemble ...The dynamin-related human guanylate-binding protein 1 (GBP1) mediates host defenses against microbial pathogens. Upon GTP binding and hydrolysis, auto-inhibited GBP1 monomers dimerize and assemble into soluble and membrane-bound oligomers, which are crucial for innate immune responses. How higher-order GBP1 oligomers are built from dimers, and how assembly is coordinated with nucleotide-dependent conformational changes, has remained elusive. Here, we present cryo-electron microscopy-based structural data of soluble and membrane-bound GBP1 oligomers, which show that GBP1 assembles in an outstretched dimeric conformation. We identify a surface-exposed helix in the large GTPase domain that contributes to the oligomerization interface, and we probe its nucleotide- and dimerization-dependent movements that facilitate the formation of an antimicrobial protein coat on a gram-negative bacterial pathogen. Our results reveal a sophisticated activation mechanism for GBP1, in which nucleotide-dependent structural changes coordinate dimerization, oligomerization, and membrane binding to allow encapsulation of pathogens within an antimicrobial protein coat. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18806.map.gz emd_18806.map.gz | 1.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18806-v30.xml emd-18806-v30.xml emd-18806.xml emd-18806.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18806_fsc.xml emd_18806_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
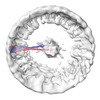
| Images |  emd_18806.png emd_18806.png | 92.8 KB | ||
| Masks |  emd_18806_msk_1.map emd_18806_msk_1.map | 8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18806.cif.gz emd-18806.cif.gz | 6.1 KB | ||
| Others |  emd_18806_half_map_1.map.gz emd_18806_half_map_1.map.gz emd_18806_half_map_2.map.gz emd_18806_half_map_2.map.gz | 7.4 MB 7.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18806 http://ftp.pdbj.org/pub/emdb/structures/EMD-18806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18806 | HTTPS FTP |
-Related structure data
| Related structure data |  8r1aMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18806.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18806.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 4.4 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18806_msk_1.map emd_18806_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18806_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18806_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Membrane-bound oligomer of human guanylate-binding protein 1 (GBP1)
| Entire | Name: Membrane-bound oligomer of human guanylate-binding protein 1 (GBP1) |
|---|---|
| Components |
|
-Supramolecule #1: Membrane-bound oligomer of human guanylate-binding protein 1 (GBP1)
| Supramolecule | Name: Membrane-bound oligomer of human guanylate-binding protein 1 (GBP1) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: in complex with GDP-AlFx |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanylate binding protein 1
| Macromolecule | Name: Guanylate binding protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 69.26793 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MRGSHHHHHH GSASEIHMTG PMCLIENTNG RLMANPEALK ILSAITQPMV VVAIVGLYRT GKSYLMNKLA GKKKGFSLGS TVQSHTKGI WMWCVPHPKK PGHILVLLDT EGLGDVEKGD NQNDSWIFAL AVLLSSTFVY NSIGTINQQA MDQLYYVTEL T HRIRSKSS ...String: MRGSHHHHHH GSASEIHMTG PMCLIENTNG RLMANPEALK ILSAITQPMV VVAIVGLYRT GKSYLMNKLA GKKKGFSLGS TVQSHTKGI WMWCVPHPKK PGHILVLLDT EGLGDVEKGD NQNDSWIFAL AVLLSSTFVY NSIGTINQQA MDQLYYVTEL T HRIRSKSS PDENENEVED SADFVSFFPD FVWTLRDFSL DLEADGQPLT PDEYLTYSLK LKKGTSQKDE TFNLPRLCIR KF FPKKKCF VFDRPVHRRK LAQLEKLQDE ELDPEFVQQV ADFCSYIFSN SKTKTLSGGI QVNGPRLESL VLTYVNAISS GDL PCMENA VLALAQIENS AAVQKAIAHY EQQMGQKVQL PTESLQELLD LHRDSEREAI EVFIRSSFKD VDHLFQKELA AQLE KKRDD FCKQNQEASS DRCSGLLQVI FSPLEEEVKA GIYSKPGGYR LFVQKLQDLK KKYYEEPRKG IQAEEILQTY LKSKE SMTD AILQTDQTLT EKEKEIEVER VKAESAQASA KMLQEMQRKN EQMMEQKERS YQEHLKQLTE KMENDRVQLL KEQERT LAL KLQEQEQLLK EGFQKESRIM KNEIQDLQTK MRRRKACTIS UniProtKB: Guanylate binding protein 1 |
-Macromolecule #2: ALUMINUM FLUORIDE
| Macromolecule | Name: ALUMINUM FLUORIDE / type: ligand / ID: 2 / Number of copies: 6 / Formula: AF3 |
|---|---|
| Molecular weight | Theoretical: 83.977 Da |
| Chemical component information |  ChemComp-AF3: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 6 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.68 mg/mL |
|---|---|
| Buffer | pH: 7.9 Details: 50 mM Tris-HCl, 150 mM NaCl, 5 mM MgCl2 (supplemented with 200 uM GDP, 300 uM AlCl3, 10 mM NaF) |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 42000 Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 42000 |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 2.8 e/Å2 Details: Tilt series were acquired with a Hybrid-STA (Sanchez et al. 2020, Nat Commun): exposure dose of 2.8 e-/A2 for non-zero tilted projection and 14.4 e-/A2 for zero tilted projection |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z
Z Y
Y X
X


























