[English] 日本語
 Yorodumi
Yorodumi- EMDB-18386: Archaeoglobus fulgidus AfAgo complex with AfAgo-N protein (fAfAgo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 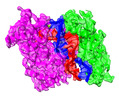 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Archaeoglobus fulgidus AfAgo complex with AfAgo-N protein (fAfAgo) bound with 17 nt RNA guide and 17 nt DNA target | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  ARGONAUTE / ARGONAUTE /  PIWI DOMAIN / PIWI DOMAIN /  PROTEIN-DNA COMPLEX / PROTEIN-DNA COMPLEX /  DNA BINDING PROTEIN DNA BINDING PROTEIN | |||||||||
| Function / homology |  Piwi domain / Piwi domain profile. / Piwi domain / Piwi domain profile. /  Piwi domain / Piwi domain /  Piwi / Ribonuclease H superfamily / Piwi / Ribonuclease H superfamily /  nucleic acid binding / Ribonuclease H-like superfamily / Uncharacterized protein / nucleic acid binding / Ribonuclease H-like superfamily / Uncharacterized protein /  Piwi protein Piwi protein Function and homology information Function and homology information | |||||||||
| Biological species |    Archaeoglobus fulgidus DSM 4304 (archaea) / Archaeoglobus fulgidus DSM 4304 (archaea) /   Escherichia coli (E. coli) / Escherichia coli (E. coli) /    Archaeoglobus fulgidus (archaea) Archaeoglobus fulgidus (archaea) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.43 Å cryo EM / Resolution: 3.43 Å | |||||||||
 Authors Authors | Manakova EN / Zaremba M / Pocevicuite R / Golovinas E / Zagorskaite E / Silanskas A | |||||||||
| Funding support | Lithuania, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein. Authors: Elena Manakova / Edvardas Golovinas / Reda Pocevičiūtė / Giedrius Sasnauskas / Arunas Silanskas / Danielis Rutkauskas / Marija Jankunec / Evelina Zagorskaitė / Edvinas Jurgelaitis / ...Authors: Elena Manakova / Edvardas Golovinas / Reda Pocevičiūtė / Giedrius Sasnauskas / Arunas Silanskas / Danielis Rutkauskas / Marija Jankunec / Evelina Zagorskaitė / Edvinas Jurgelaitis / Algirdas Grybauskas / Česlovas Venclovas / Mindaugas Zaremba Abstract: Argonaute (Ago) proteins are present in all three domains of life (bacteria, archaea and eukaryotes). They use small (15-30 nucleotides) oligonucleotide guides to bind complementary nucleic acid ...Argonaute (Ago) proteins are present in all three domains of life (bacteria, archaea and eukaryotes). They use small (15-30 nucleotides) oligonucleotide guides to bind complementary nucleic acid targets and are responsible for gene expression regulation, mobile genome element silencing, and defence against viruses or plasmids. According to their domain organization, Agos are divided into long and short Agos. Long Agos found in prokaryotes (long-A and long-B pAgos) and eukaryotes (eAgos) comprise four major functional domains (N, PAZ, MID and PIWI) and two structural linker domains L1 and L2. The majority (∼60%) of pAgos are short pAgos, containing only the MID and inactive PIWI domains. Here we focus on the prokaryotic Argonaute AfAgo from Archaeoglobus fulgidus DSM4304. Although phylogenetically classified as a long-B pAgo, AfAgo contains only MID and catalytically inactive PIWI domains, akin to short pAgos. We show that AfAgo forms a heterodimeric complex with a protein encoded upstream in the same operon, which is a structural equivalent of the N-L1-L2 domains of long pAgos. This complex, structurally equivalent to a long PAZ-less pAgo, outperforms standalone AfAgo in guide RNA-mediated target DNA binding. Our findings provide a missing piece to one of the first and the most studied pAgos. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18386.map.gz emd_18386.map.gz | 24.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18386-v30.xml emd-18386-v30.xml emd-18386.xml emd-18386.xml | 26.2 KB 26.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18386_fsc.xml emd_18386_fsc.xml | 6.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_18386.png emd_18386.png | 164.5 KB | ||
| Filedesc metadata |  emd-18386.cif.gz emd-18386.cif.gz | 7.4 KB | ||
| Others |  emd_18386_half_map_1.map.gz emd_18386_half_map_1.map.gz emd_18386_half_map_2.map.gz emd_18386_half_map_2.map.gz | 25.1 MB 25.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18386 http://ftp.pdbj.org/pub/emdb/structures/EMD-18386 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18386 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18386 | HTTPS FTP |
-Related structure data
| Related structure data |  8qg0MC  8ok9C  8oldC  8oljC  8pvvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18386.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18386.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_18386_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18386_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Heterodimeric AfAgo and AfAgo-N complex with 17 nt RNA guide and ...
| Entire | Name: Heterodimeric AfAgo and AfAgo-N complex with 17 nt RNA guide and 17 nt DNA target |
|---|---|
| Components |
|
-Supramolecule #1: Heterodimeric AfAgo and AfAgo-N complex with 17 nt RNA guide and ...
| Supramolecule | Name: Heterodimeric AfAgo and AfAgo-N complex with 17 nt RNA guide and 17 nt DNA target type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 100 KDa |
-Supramolecule #2: AfAgo
| Supramolecule | Name: AfAgo / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:    Archaeoglobus fulgidus DSM 4304 (archaea) Archaeoglobus fulgidus DSM 4304 (archaea) |
-Supramolecule #3: AfAgo-N
| Supramolecule | Name: AfAgo-N / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:    Archaeoglobus fulgidus DSM 4304 (archaea) Archaeoglobus fulgidus DSM 4304 (archaea) |
-Supramolecule #4: DNA target oligonucleotide
| Supramolecule | Name: DNA target oligonucleotide / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Supramolecule #5: 5'p-RNA guide
| Supramolecule | Name: 5'p-RNA guide / type: complex / ID: 5 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: Piwi protein
| Macromolecule | Name: Piwi protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Archaeoglobus fulgidus (archaea) / Strain: DSM4304 Archaeoglobus fulgidus (archaea) / Strain: DSM4304 |
| Molecular weight | Theoretical: 49.302434 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MMEYKIVENG LTYRIGNGAS VPISNTGELI KGLRNYGPYE VPSLKYNQIA LIHNNQFSSL INQLKSQISS KIDEVWHIHN INISEFIYD SPHFDSIKSQ VDNAIDTGVD GIMLVLPEYN TPLYYKLKSY LINSIPSQFM RYDILSNRNL TFYVDNLLVQ F VSKLGGKP ...String: MMEYKIVENG LTYRIGNGAS VPISNTGELI KGLRNYGPYE VPSLKYNQIA LIHNNQFSSL INQLKSQISS KIDEVWHIHN INISEFIYD SPHFDSIKSQ VDNAIDTGVD GIMLVLPEYN TPLYYKLKSY LINSIPSQFM RYDILSNRNL TFYVDNLLVQ F VSKLGGKP WILNVDPEKG SDIIIGTGAT RIDNVNLFCF AMVFKKDGTM LWNEISPIVT SSEYLTYLKS TIKKVVYGFK KS NPDWDVE KLTLHVSGKR PKMKDGETKI LKETVEELKK QEMVSRDVKY AILHLNETHP FWVMGDPNNR FHPYEGTKVK LSS KRYLLT LLQPYLKRNG LEMVTPIKPL SVEIVSDNWT SEEYYHNVHE ILDEIYYLSK MNWRGFRSRN LPVTVNYPKL VAGI IANVN RYGGYPINPE GNRSLQTNPW FL UniProtKB:  Piwi protein Piwi protein |
-Macromolecule #2: AfAgo-N protein
| Macromolecule | Name: AfAgo-N protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Archaeoglobus fulgidus (archaea) / Strain: DSM8774 Archaeoglobus fulgidus (archaea) / Strain: DSM8774 |
| Molecular weight | Theoretical: 31.356576 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MGGSHHHHHH GMASENLYFQ GGGGEIPLSS GNVNTPDVRS SGILYINIYP IVNYPETIKV SAIPYYEEFL PGKWKKRIGD LIYLYGYGI ENEFDEIDNS NALFGKIFRK YLLDILSENI ATPWQLKELG STLRLVKEIT ENYEFSNIIK LQYELIINVH H WQNTNFGI ...String: MGGSHHHHHH GMASENLYFQ GGGGEIPLSS GNVNTPDVRS SGILYINIYP IVNYPETIKV SAIPYYEEFL PGKWKKRIGD LIYLYGYGI ENEFDEIDNS NALFGKIFRK YLLDILSENI ATPWQLKELG STLRLVKEIT ENYEFSNIIK LQYELIINVH H WQNTNFGI IVDLKINILD RENNQRISYT KIKDKYGESV KKKIWVSVQA FHRHLTPEGK KYATAMRDKF NLLTGLLKEA FG SSEDEKT FSTPDGEIKI VFKPLEIVEV SNNDGI UniProtKB: Uncharacterized protein |
-Macromolecule #3: DNA target 17 nt
| Macromolecule | Name: DNA target 17 nt / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 5.202384 KDa |
| Sequence | String: (DA)(DT)(DT)(DC)(DG)(DG)(DC)(DC)(DG)(DT) (DG)(DT)(DA)(DC)(DA)(DA)(DT) |
-Macromolecule #4: RNA guide 17 nt
| Macromolecule | Name: RNA guide 17 nt / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 5.427286 KDa |
| Sequence | String: AUUGUACACG GCCGAAU |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | fAfAgo complex with 17/17 guide-target heteroduplexes was mixed and applied on grid |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 92000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 92000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Number grids imaged: 1 / Number real images: 2152 / Average exposure time: 46.33 sec. / Average electron dose: 31.0 e/Å2 |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 48 | |||||||||
| Output model |  PDB-8qg0: |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X