[English] 日本語
 Yorodumi
Yorodumi- EMDB-17790: Structure of tissue-specific lipid scramblase ATG9B homotrimer, r... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of tissue-specific lipid scramblase ATG9B homotrimer, refined without imposing symmetry | ||||||||||||||||||
 Map data Map data | ATG9B C1 map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords |  membrane protein / lipid scramblase / membrane protein / lipid scramblase /  autophagy / phagopore / lipid transporter / Atg9 / Atg9B / LIPID TRANSPORT autophagy / phagopore / lipid transporter / Atg9 / Atg9B / LIPID TRANSPORT | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information phospholipid scramblase activity / programmed necrotic cell death / nucleophagy / protein localization to phagophore assembly site / phagophore assembly site membrane / phagophore assembly site / bone morphogenesis / phospholipid scramblase activity / programmed necrotic cell death / nucleophagy / protein localization to phagophore assembly site / phagophore assembly site membrane / phagophore assembly site / bone morphogenesis /  Macroautophagy / Macroautophagy /  autophagosome assembly / autophagosome assembly /  autophagosome ... autophagosome ... phospholipid scramblase activity / programmed necrotic cell death / nucleophagy / protein localization to phagophore assembly site / phagophore assembly site membrane / phagophore assembly site / bone morphogenesis / phospholipid scramblase activity / programmed necrotic cell death / nucleophagy / protein localization to phagophore assembly site / phagophore assembly site membrane / phagophore assembly site / bone morphogenesis /  Macroautophagy / Macroautophagy /  autophagosome assembly / autophagosome assembly /  autophagosome / autophagosome /  trans-Golgi network / recycling endosome membrane / trans-Golgi network / recycling endosome membrane /  Golgi membrane / endoplasmic reticulum membrane Golgi membrane / endoplasmic reticulum membraneSimilarity search - Function | ||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
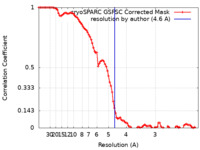
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.6 Å cryo EM / Resolution: 4.6 Å | ||||||||||||||||||
 Authors Authors | Chiduza GN / Pye VE / Tooze SA / Cherepanov P | ||||||||||||||||||
| Funding support |  United Kingdom, European Union, 5 items United Kingdom, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Autophagy / Year: 2024 Journal: Autophagy / Year: 2024Title: ATG9B is a tissue-specific homotrimeric lipid scramblase that can compensate for ATG9A. Authors: George N Chiduza / Acely Garza-Garcia / Eugenia Almacellas / Stefano De Tito / Valerie E Pye / Alexander R van Vliet / Peter Cherepanov / Sharon A Tooze /  Abstract: Macroautophagy/autophagy is a fundamental aspect of eukaryotic biology, and the autophay-related protein ATG9A is part of the core machinery facilitating this process. In addition to ATG9A ...Macroautophagy/autophagy is a fundamental aspect of eukaryotic biology, and the autophay-related protein ATG9A is part of the core machinery facilitating this process. In addition to ATG9A vertebrates encode ATG9B, a poorly characterized paralog expressed in a subset of tissues. Herein, we characterize the structure of human ATG9B revealing the conserved homotrimeric quaternary structure and explore the conformational dynamics of the protein. Consistent with the experimental structure and computational chemistry, we establish that ATG9B is a functional lipid scramblase. We show that ATG9B can compensate for the absence of ATG9A in starvation-induced autophagy displaying similar subcellular trafficking and steady-state localization. Finally, we demonstrate that ATG9B can form a heteromeric complex with ATG2A. By establishing the molecular structure and function of ATG9B, our results inform the exploration of niche roles for autophagy machinery in more complex eukaryotes and reveal insights relevant across species. ATG: autophagy related; CHS: cholesteryl hemisuccinate; cryo-EM: single-particle cryogenic electron microscopy; CTF: contrast transfer function: CTH: C- terminal α helix; FSC: fourier shell correlation; HDIR: HORMA domain interacting region; LMNG: lauryl maltose neopentyl glycol; MD: molecular dynamics simulations; MSA: multiple sequence alignment; NBD-PE: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl ammonium salt); POPC: palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; RBG: repeating beta groove domain; RMSD: root mean square deviation; SEC: size-exclusion chromatography; TMH: transmembrane helix. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17790.map.gz emd_17790.map.gz | 78.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17790-v30.xml emd-17790-v30.xml emd-17790.xml emd-17790.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17790_fsc.xml emd_17790_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_17790.png emd_17790.png | 86.9 KB | ||
| Masks |  emd_17790_msk_1.map emd_17790_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17790.cif.gz emd-17790.cif.gz | 5.9 KB | ||
| Others |  emd_17790_additional_1.map.gz emd_17790_additional_1.map.gz emd_17790_half_map_1.map.gz emd_17790_half_map_1.map.gz emd_17790_half_map_2.map.gz emd_17790_half_map_2.map.gz | 74 MB 77.7 MB 77.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17790 http://ftp.pdbj.org/pub/emdb/structures/EMD-17790 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17790 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17790 | HTTPS FTP |
-Related structure data
| Related structure data |  8poeC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17790.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17790.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ATG9B C1 map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17790_msk_1.map emd_17790_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
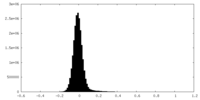
| Density Histograms |
-Additional map: ATG9B C1 DeepEMhancer map
| File | emd_17790_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ATG9B C1 DeepEMhancer map | ||||||||||||
| Projections & Slices |
| ||||||||||||
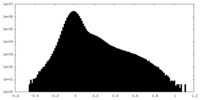
| Density Histograms |
-Half map: ATG9B C1 half map 1
| File | emd_17790_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ATG9B C1 half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
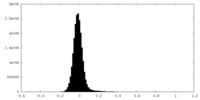
| Density Histograms |
-Half map: ATG9B C1 half map 2
| File | emd_17790_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ATG9B C1 half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
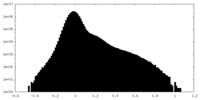
| Density Histograms |
- Sample components
Sample components
-Entire : Homotrimer of ATG9B
| Entire | Name: Homotrimer of ATG9B |
|---|---|
| Components |
|
-Supramolecule #1: Homotrimer of ATG9B
| Supramolecule | Name: Homotrimer of ATG9B / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 303 KDa |
-Macromolecule #1: Autophagy-related protein 9B
| Macromolecule | Name: Autophagy-related protein 9B / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDKDY KDDDDKDYKD DDDKHHHHHH ENLYFQMVSR MGWGGRRRRL GRWGDLGPGS VPLLPMPLPP PPPPSCRGPG GGRISIFSLS PAPHTR SSP SSFSPPTAGP PCSVLQGTGA SQSCHSALPI PATPPTQAQP AMTPASASPS WGSHSTP PL APATPTPSQQ ...String: DYKDDDDKDY KDDDDKDYKD DDDKHHHHHH ENLYFQMVSR MGWGGRRRRL GRWGDLGPGS VPLLPMPLPP PPPPSCRGPG GGRISIFSLS PAPHTR SSP SSFSPPTAGP PCSVLQGTGA SQSCHSALPI PATPPTQAQP AMTPASASPS WGSHSTP PL APATPTPSQQ CPQDSPGLRV GPLIPEQDYE RLEDCDPEGS QDSPIHGEEQ QPLLHVPE G LRGSWHHIQN LDSFFTKIYS YHQRNGFACI LLEDVFQLGQ FIFIVTFTTF LLRCVDYNV LFANQPSNHT RPGPFHSKVT LSDAILPSAQ CAERIRSSPL LVLLLVLAAG FWLVQLLRSV CNLFSYWDI QVFYREALHI PPEELSSVPW AEVQSRLLAL QRSGGLCVQP RPLTELDIHH R ILRYTNYQ VALANKGLLP ARCPLPWGGS AAFLSRGLAL NVDLLLFRGP FSLFRGGWEL PH AYKRSDQ RGALAARWGR TVLLLAALNL ALSPLVLAWQ VLHVFYSHVE LLRREPGALG ARG WSRLAR LQLRHFNELP HELRARLARA YRPAAAFLRT AAPPAPLRTL LARQLVFFAG ALFA ALLVL TVYDEDVLAV EHVLTAMTAL GVTATVARSF IPEEQCQGRA PQLLLQTALA HMHYL PEEP GPGGRDRAYR QMAQLLQYRA VSLLEELLSP LLTPLFLLFW FRPRALEIID FFHHFT VDV AGVGDICSFA LMDVKRHGHP QWLSAGQTEA SLSQRAEDGK TELSLMRFSL AHPLWRP PG HSSKFLGHLW GRVQQDAAAW GATSARGPST PGVLSNCTSP LPEAFLANLF VHPLLPPR D LSPTAPCPAA ATASLLASIS RIAQDPSSVS PGGTGGQKLA QLPELASAEM SLHVIYLHQ LHQQQQQQEP WGEAAASILS RPCSSPSQPP SPDEEKPSWS SDGSSPASSP RQQWGTQKAR NLFPGGFQV TTDTQKEPDR ASCTD UniProtKB: Autophagy-related protein 9B |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 8.5 Details: 20 mM Tris-HCl, pH 8.5; 200 mM NaCl; 1 mM TCEP; 10% glycerol supplemented with 0.001% w/v lauryl maltose neopentyl glycol and 0.0002% w/v cholesteryl hemisuccinate |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 46296 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 2 / Number real images: 37217 / Average exposure time: 1.2 sec. / Average electron dose: 48.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)