+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lipidic amyloid-beta(1-40) fibril - polymorph L3 | ||||||||||||
 Map data Map data | Lipidic amyloid-beta(1-40) fibril - polymorph L3 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  amyloid-beta / amyloid-beta /  fibril / fibril /  lipids / PROTEIN FIBRIL lipids / PROTEIN FIBRIL | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationGolgi-associated vesicle /  clathrin-coated pit / clathrin-coated pit /  heparin binding / heparin binding /  growth cone / growth cone /  perikaryon / perikaryon /  early endosome / early endosome /  Golgi apparatus / Golgi apparatus /  cell surface / cell surface /  endoplasmic reticulum / extracellular region / endoplasmic reticulum / extracellular region /  nucleus nucleusSimilarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
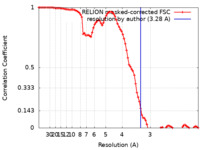
| Method | helical reconstruction /  cryo EM / Resolution: 3.28 Å cryo EM / Resolution: 3.28 Å | ||||||||||||
 Authors Authors | Frieg B / Han M / Giller K / Dienemann C / Riedel D / Becker S / Andreas LB / Griesinger C / Schroeder GF | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structures of lipidic fibrils of amyloid-β (1-40). Authors: Benedikt Frieg / Mookyoung Han / Karin Giller / Christian Dienemann / Dietmar Riedel / Stefan Becker / Loren B Andreas / Christian Griesinger / Gunnar F Schröder /  Abstract: Alzheimer's disease (AD) is a progressive and incurable neurodegenerative disease characterized by the extracellular deposition of amyloid plaques. Investigation into the composition of these plaques ...Alzheimer's disease (AD) is a progressive and incurable neurodegenerative disease characterized by the extracellular deposition of amyloid plaques. Investigation into the composition of these plaques revealed a high amount of amyloid-β (Aβ) fibrils and a high concentration of lipids, suggesting that fibril-lipid interactions may also be relevant for the pathogenesis of AD. Therefore, we grew Aβ40 fibrils in the presence of lipid vesicles and determined their structure by cryo-electron microscopy (cryo-EM) to high resolution. The fold of the major polymorph is similar to the structure of brain-seeded fibrils reported previously. The majority of the lipids are bound to the fibrils, as we show by cryo-EM and NMR spectroscopy. This apparent lipid extraction from vesicles observed here in vitro provides structural insights into potentially disease-relevant fibril-lipid interactions. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17234.map.gz emd_17234.map.gz | 5.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17234-v30.xml emd-17234-v30.xml emd-17234.xml emd-17234.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17234_fsc.xml emd_17234_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_17234.png emd_17234.png | 27.4 KB | ||
| Filedesc metadata |  emd-17234.cif.gz emd-17234.cif.gz | 5.2 KB | ||
| Others |  emd_17234_half_map_1.map.gz emd_17234_half_map_1.map.gz emd_17234_half_map_2.map.gz emd_17234_half_map_2.map.gz | 46 MB 45.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17234 http://ftp.pdbj.org/pub/emdb/structures/EMD-17234 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17234 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17234 | HTTPS FTP |
-Related structure data
| Related structure data |  8owdMC  8ovkC  8ovmC  8oweC  8owjC  8owkC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17234.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17234.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Lipidic amyloid-beta(1-40) fibril - polymorph L3 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
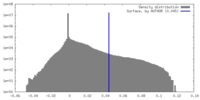
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Lipidic amyloid-beta(1-40) fibril - polymorph L3 (half map 1)
| File | emd_17234_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Lipidic amyloid-beta(1-40) fibril - polymorph L3 (half map 1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
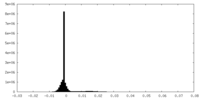
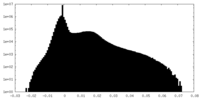
| Density Histograms |
-Half map: Lipidic amyloid-beta(1-40) fibril - polymorph L3 (half map 2)
| File | emd_17234_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Lipidic amyloid-beta(1-40) fibril - polymorph L3 (half map 2) | ||||||||||||
| Projections & Slices |
| ||||||||||||
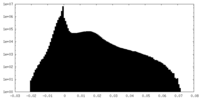
| Density Histograms |
- Sample components
Sample components
-Entire : The L3 amyloid-beta(1-40) fibril in complex with lipids
| Entire | Name: The L3 amyloid-beta(1-40) fibril in complex with lipids |
|---|---|
| Components |
|
-Supramolecule #1: The L3 amyloid-beta(1-40) fibril in complex with lipids
| Supramolecule | Name: The L3 amyloid-beta(1-40) fibril in complex with lipids type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Amyloid-beta A4 protein
| Macromolecule | Name: Amyloid-beta A4 protein / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.335852 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: DAEFRHDSGY EVHHQKLVFF AEDVGSNKGA IIGLMVGGVV UniProtKB: Amyloid-beta A4 protein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)