+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Urea Transporter UT-A (N-Terminal Domain Model) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SLC14A2 / UT2 / UT-A /  Urea Transporter / Inhibitor / Solute Carrier / Urea Transporter / Inhibitor / Solute Carrier /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationurea transport / Transport of bile salts and organic acids, metal ions and amine compounds / urea transmembrane transporter activity / urea transmembrane transport /  cell adhesion molecule binding / transmembrane transport / apical plasma membrane / cell adhesion molecule binding / transmembrane transport / apical plasma membrane /  membrane / membrane /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
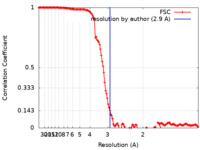
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.9 Å cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Chi G / Pike ACW / Maclean EM / Mukhopadhyay SMM / Bohstedt T / Scacioc A / Wang D / McKinley G / Fernandez-Cid A / Arrowsmith CH ...Chi G / Pike ACW / Maclean EM / Mukhopadhyay SMM / Bohstedt T / Scacioc A / Wang D / McKinley G / Fernandez-Cid A / Arrowsmith CH / Bountra C / Edwards A / Burgess-Brown NA / van Putte W / Duerr K | |||||||||
| Funding support |  United Kingdom, European Union, 2 items United Kingdom, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structural characterization of human urea transporters UT-A and UT-B and their inhibition. Authors: Gamma Chi / Larissa Dietz / Haiping Tang / Matthew Snee / Andreea Scacioc / Dong Wang / Gavin Mckinley / Shubhashish M M Mukhopadhyay / Ashley C W Pike / Rod Chalk / Nicola A Burgess-Brown / ...Authors: Gamma Chi / Larissa Dietz / Haiping Tang / Matthew Snee / Andreea Scacioc / Dong Wang / Gavin Mckinley / Shubhashish M M Mukhopadhyay / Ashley C W Pike / Rod Chalk / Nicola A Burgess-Brown / Jean-Pierre Timmermans / Wouter van Putte / Carol V Robinson / Katharina L Dürr /   Abstract: In this study, we present the structures of human urea transporters UT-A and UT-B to characterize them at molecular level and to detail the mechanism of UT-B inhibition by its selective inhibitor, ...In this study, we present the structures of human urea transporters UT-A and UT-B to characterize them at molecular level and to detail the mechanism of UT-B inhibition by its selective inhibitor, UTB-14. High-resolution structures of both transporters establish the structural basis for the inhibitor's selectivity to UT-B, and the identification of multiple binding sites for the inhibitor will aid with the development of drug lead molecules targeting both transporters. Our study also discovers phospholipids associating with the urea transporters by combining structural observations, native MS, and lipidomics analysis. These insights improve our understanding of urea transporter function at a molecular level and provide a blueprint for a structure-guided design of therapeutics targeting these transporters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16110.map.gz emd_16110.map.gz | 97 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16110-v30.xml emd-16110-v30.xml emd-16110.xml emd-16110.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16110_fsc.xml emd_16110_fsc.xml | 15.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_16110.png emd_16110.png | 216.7 KB | ||
| Masks |  emd_16110_msk_1.map emd_16110_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16110.cif.gz emd-16110.cif.gz | 6.7 KB | ||
| Others |  emd_16110_half_map_1.map.gz emd_16110_half_map_1.map.gz emd_16110_half_map_2.map.gz emd_16110_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16110 http://ftp.pdbj.org/pub/emdb/structures/EMD-16110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16110 | HTTPS FTP |
-Related structure data
| Related structure data |  8bloMC  8blpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16110.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16110.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16110_msk_1.map emd_16110_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16110_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16110_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Trimer-like complex of Human UT-A
| Entire | Name: Trimer-like complex of Human UT-A |
|---|---|
| Components |
|
-Supramolecule #1: Trimer-like complex of Human UT-A
| Supramolecule | Name: Trimer-like complex of Human UT-A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: Urea transporter 2
| Macromolecule | Name: Urea transporter 2 / type: protein_or_peptide / ID: 1 Details: This model is based on the N-terminal half of the protein UT-A. However, UT-A consists of two similar domains, and the map is sometimes ambiguous whether the structure is of the N-terminal ...Details: This model is based on the N-terminal half of the protein UT-A. However, UT-A consists of two similar domains, and the map is sometimes ambiguous whether the structure is of the N-terminal or C-terminal half. Much of the map is in line with amino acids from its N-terminal domain. Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 100.124172 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSDPHSSPLL PEPLSSRYKL YEAEFTSPSW PSTSPDTHPA LPLLEMPEEK DLRSSNEDSH IVKIEKLNER SKRKDDGVAH RDSAGQRCI CLSKAVGYLT GDMKEYRIWL KDKHLALQFI DWVLRGTAQV MFINNPLSGL IIFIGLLIQN PWWTITGGLG T VVSTLTAL ...String: MSDPHSSPLL PEPLSSRYKL YEAEFTSPSW PSTSPDTHPA LPLLEMPEEK DLRSSNEDSH IVKIEKLNER SKRKDDGVAH RDSAGQRCI CLSKAVGYLT GDMKEYRIWL KDKHLALQFI DWVLRGTAQV MFINNPLSGL IIFIGLLIQN PWWTITGGLG T VVSTLTAL ALGQDRSAIA SGLHGYNGML VGLLMAVFSE KLDYYWWLLF PVTFTAMSCP VLSSALNSIF SKWDLPVFTL PF NIAVTLY LAATGHYNLF FPTTLVEPVS SVPNITWTEM EMPLLLQAIP VGVGQVYGCD NPWTGGVFLV ALFISSPLIC LHA AIGSIV GLLAALSVAT PFETIYTGLW SYNCVLSCIA IGGMFYALTW QTHLLALICA LFCAYMEAAI SNIMSVVGVP PGTW AFCLA TIIFLLLTTN NPAIFRLPLS KVTYPEANRI YYLTVKSGEE EKAPSGGGGE HPPTAGPKVE EGSEAVLSKH RSVFH IEWS SIRRRSKVFG KGEHQERQNK DPFPYRYRKP TVELLDLDTM EESSEIKVET NISKTSWIRS SMAASGKRVS KALSYI TGE MKECGEGLKD KSPVFQFFDW VLRGTSQVMF VNNPLSGILI ILGLFIQNPW WAISGCLGTI MSTLTALILS QDKSAIA AG FHGYNGVLVG LLMAVFSDKG DYYWWLLLPV IIMSMSCPIL SSALGTIFSK WDLPVFTLPF NITVTLYLAA TGHYNLFF P TTLLQPASAM PNITWSEVQV PLLLRAIPVG IGQVYGCDNP WTGGIFLIAL FISSPLICLH AAIGSTMGML AALTIATPF DSIYFGLCGF NSTLACIAIG GMFYVITWQT HLLAIACALF AAYLGAALAN MLSVFGLPPC TWPFCLSALT FLLLTTNNPA IYKLPLSKV TYPEANRIYY LSQEAENLYF Q UniProtKB:  Urea transporter 2 Urea transporter 2 |
-Macromolecule #2: di-heneicosanoyl phosphatidyl choline
| Macromolecule | Name: di-heneicosanoyl phosphatidyl choline / type: ligand / ID: 2 / Number of copies: 9 / Formula: PLD |
|---|---|
| Molecular weight | Theoretical: 875.313 Da |
| Chemical component information |  ChemComp-PLD: |
-Macromolecule #3: Lauryl Maltose Neopentyl Glycol
| Macromolecule | Name: Lauryl Maltose Neopentyl Glycol / type: ligand / ID: 3 / Number of copies: 15 / Formula: LMN |
|---|---|
| Molecular weight | Theoretical: 1.005188 KDa |
| Chemical component information |  ChemComp-AV0: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 18 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Homemade / Support film - Material: GRAPHENE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 52.63 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)