[English] 日本語
 Yorodumi
Yorodumi- EMDB-15947: Direct observation of the open conformational state of formin mDi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Direct observation of the open conformational state of formin mDia1 at actin filament barbed end | |||||||||
 Map data Map data | Actin barbed-end - Formin "Open State" | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Actin binding protein /  Complex / barbed-end / Complex / barbed-end /  formin / formin /  MOTOR PROTEIN MOTOR PROTEIN | |||||||||
| Biological species |   Mus musculus (house mouse) / Mus musculus (house mouse) /   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) | |||||||||
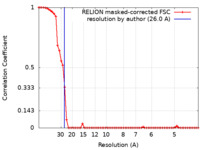
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 26.0 Å negative staining / Resolution: 26.0 Å | |||||||||
 Authors Authors | Maufront J / Guichard B / Cao L / Di Cicco A / Jegou A / Romet-Lemonne G / Bertin A | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Biol Cell / Year: 2023 Journal: Mol Biol Cell / Year: 2023Title: Direct observation of the conformational states of formin mDia1 at actin filament barbed ends and along the filament. Authors: Julien Maufront / Bérengère Guichard / Lu-Yan Cao / Aurélie Di Cicco / Antoine Jégou / Guillaume Romet-Lemonne / Aurélie Bertin /  Abstract: The fine regulation of actin polymerization is essential to control cell motility and architecture and to perform essential cellular functions. Formins are key regulators of actin filament assembly, ...The fine regulation of actin polymerization is essential to control cell motility and architecture and to perform essential cellular functions. Formins are key regulators of actin filament assembly, known to processively elongate filament barbed ends and increase their polymerization rate. Different models have been extrapolated to describe the molecular mechanism governing the processive motion of formin FH2 domains at polymerizing barbed ends. Using negative stain electron microscopy, we directly identified for the first time two conformations of the mDia1 formin FH2 domains in interaction with the barbed ends of actin filaments. These conformations agree with the speculated open and closed conformations of the "stair-stepping" model. We observed the FH2 dimers to be in the open conformation for 79% of the data, interacting with the two terminal actin subunits of the barbed end while they interact with three actin subunits in the closed conformation. In addition, we identified and characterized the structure of single FH2 dimers encircling the core of actin filaments, and reveal their ability to spontaneously depart from barbed ends. #1:  Journal: BiorXiv / Year: 2022 Journal: BiorXiv / Year: 2022Title: Direct observation of the open conformational state of formin mDia1 at actin filament barbed end Authors: Maufront J / Bertin A | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15947.map.gz emd_15947.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15947-v30.xml emd-15947-v30.xml emd-15947.xml emd-15947.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15947_fsc.xml emd_15947_fsc.xml | 6 KB | Display |  FSC data file FSC data file |
| Images |  emd_15947.png emd_15947.png | 39.3 KB | ||
| Masks |  emd_15947_msk_1.map emd_15947_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15947.cif.gz emd-15947.cif.gz | 4.4 KB | ||
| Others |  emd_15947_half_map_1.map.gz emd_15947_half_map_1.map.gz emd_15947_half_map_2.map.gz emd_15947_half_map_2.map.gz | 17 MB 17 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15947 http://ftp.pdbj.org/pub/emdb/structures/EMD-15947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15947 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15947.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15947.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Actin barbed-end - Formin "Open State" | ||||||||||||||||||||
| Voxel size | X=Y=Z: 2.13 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15947_msk_1.map emd_15947_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Actin barbed-end - Formin "Open State"
| File | emd_15947_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Actin barbed-end - Formin "Open State" | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Actin barbed-end - Formin "Open State"
| File | emd_15947_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Actin barbed-end - Formin "Open State" | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Actin barbed-end bound by formin mDia1 dimer in his "open state"
| Entire | Name: Actin barbed-end bound by formin mDia1 dimer in his "open state" |
|---|---|
| Components |
|
-Supramolecule #1: Actin barbed-end bound by formin mDia1 dimer in his "open state"
| Supramolecule | Name: Actin barbed-end bound by formin mDia1 dimer in his "open state" type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: Formin mDia1 dimer
| Supramolecule | Name: Formin mDia1 dimer / type: organelle_or_cellular_component / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Supramolecule #3: Alpha Actin subunits assembled in helicoidal F-actin filament
| Supramolecule | Name: Alpha Actin subunits assembled in helicoidal F-actin filament type: organelle_or_cellular_component / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation #1
Sample preparation #1
| Preparation ID | 1 |
|---|---|
| Concentration | 0.04 mg/mL |
| Buffer | pH: 7.8 |
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
- Sample preparation #2
Sample preparation #2
| Preparation ID | 2 |
|---|---|
| Concentration | 0.01 mg/mL |
| Buffer | pH: 7.8 |
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 20.0 e/Å2 |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X