[English] 日本語
 Yorodumi
Yorodumi- EMDB-15561: Peripheral stalk of Trypanosoma brucei mitochondrial ATP synthase -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Peripheral stalk of Trypanosoma brucei mitochondrial ATP synthase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information kinetoplast / mitochondrial proton-transporting ATP synthase, stator stalk / mitochondrial proton-transporting ATP synthase complex / proton motive force-driven mitochondrial ATP synthesis / proton-transporting ATP synthase complex, catalytic core F(1) / proton-transporting ATP synthase activity, rotational mechanism / kinetoplast / mitochondrial proton-transporting ATP synthase, stator stalk / mitochondrial proton-transporting ATP synthase complex / proton motive force-driven mitochondrial ATP synthesis / proton-transporting ATP synthase complex, catalytic core F(1) / proton-transporting ATP synthase activity, rotational mechanism /  mitochondrial membrane / mitochondrial membrane /  mitochondrion / mitochondrion /  nucleoplasm / nucleoplasm /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Trypanosoma brucei brucei (eukaryote) Trypanosoma brucei brucei (eukaryote) | |||||||||
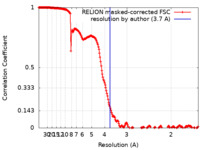
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Muehleip A / Gahura O / Zikova A / Amunts A | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases. Authors: Ondřej Gahura / Alexander Mühleip / Carolina Hierro-Yap / Brian Panicucci / Minal Jain / David Hollaus / Martina Slapničková / Alena Zíková / Alexey Amunts /   Abstract: Mitochondrial ATP synthase forms stable dimers arranged into oligomeric assemblies that generate the inner-membrane curvature essential for efficient energy conversion. Here, we report cryo-EM ...Mitochondrial ATP synthase forms stable dimers arranged into oligomeric assemblies that generate the inner-membrane curvature essential for efficient energy conversion. Here, we report cryo-EM structures of the intact ATP synthase dimer from Trypanosoma brucei in ten different rotational states. The model consists of 25 subunits, including nine lineage-specific, as well as 36 lipids. The rotary mechanism is influenced by the divergent peripheral stalk, conferring a greater conformational flexibility. Proton transfer in the lumenal half-channel occurs via a chain of five ordered water molecules. The dimerization interface is formed by subunit-g that is critical for interactions but not for the catalytic activity. Although overall dimer architecture varies among eukaryotes, we find that subunit-g together with subunit-e form an ancestral oligomerization motif, which is shared between the trypanosomal and mammalian lineages. Therefore, our data defines the subunit-g/e module as a structural component determining ATP synthase oligomeric assemblies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15561.map.gz emd_15561.map.gz | 360.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15561-v30.xml emd-15561-v30.xml emd-15561.xml emd-15561.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15561_fsc.xml emd_15561_fsc.xml | 19.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_15561.png emd_15561.png | 66.6 KB | ||
| Masks |  emd_15561_msk_1.map emd_15561_msk_1.map | 669.9 MB |  Mask map Mask map | |
| Others |  emd_15561_half_map_1.map.gz emd_15561_half_map_1.map.gz emd_15561_half_map_2.map.gz emd_15561_half_map_2.map.gz | 534.2 MB 534.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15561 http://ftp.pdbj.org/pub/emdb/structures/EMD-15561 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15561 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15561 | HTTPS FTP |
-Related structure data
| Related structure data |  8ap8MC  8ap6C  8ap7C  8ap9C  8apaC  8apbC  8apcC  8apdC  8apeC  8apfC  8apgC  8aphC  8apjC  8apkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15561.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15561.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15561_msk_1.map emd_15561_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15561_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15561_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mitochondrial ATP synthase dimer from Trypanosoma brucei
| Entire | Name: mitochondrial ATP synthase dimer from Trypanosoma brucei |
|---|---|
| Components |
|
-Supramolecule #1: mitochondrial ATP synthase dimer from Trypanosoma brucei
| Supramolecule | Name: mitochondrial ATP synthase dimer from Trypanosoma brucei type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Trypanosoma brucei brucei (eukaryote) / Strain: Lister 427 Trypanosoma brucei brucei (eukaryote) / Strain: Lister 427 |
-Macromolecule #1: OSCP
| Macromolecule | Name: OSCP / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Trypanosoma brucei brucei (eukaryote) Trypanosoma brucei brucei (eukaryote) |
| Molecular weight | Theoretical: 28.869924 KDa |
| Sequence | String: MFRRLSSSAR AVVAARFYTP PEGLKKLYAS DFENSKYPLN IVPSDSVLFA KFLYKAAEEK GNFDNILSDF QKIAAAASKL PIFWERTAV VEKIPEFKQL SEPTFFTLVW MQNNGMLELI QEVAEVYETF VNAKQKKAVA KIFVAPGGEK NVEEARRVAE E LHKGLKEL ...String: MFRRLSSSAR AVVAARFYTP PEGLKKLYAS DFENSKYPLN IVPSDSVLFA KFLYKAAEEK GNFDNILSDF QKIAAAASKL PIFWERTAV VEKIPEFKQL SEPTFFTLVW MQNNGMLELI QEVAEVYETF VNAKQKKAVA KIFVAPGGEK NVEEARRVAE E LHKGLKEL ADYTLVLKTV VDRTIVKGFA VELAGQYVNK AEGQQKQAGR ADEVDYTNLP APKPQKTVWD DNIETEVLRK YL DGLSQYD MEEAKYGV |
-Macromolecule #2: ATPTB4
| Macromolecule | Name: ATPTB4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Trypanosoma brucei brucei (eukaryote) Trypanosoma brucei brucei (eukaryote) |
| Molecular weight | Theoretical: 17.218723 KDa |
| Sequence | String: MRRTFISFSA ASAAAAAPVT STKMQTLHKL LTGEVSFKNK APVKDCNIVH QFGENWATEL SAYAKTLPAE QQKIIVRQIA RVKLTRYTV AELAAYCGDG PALLDETARA ANIEQGVAFV KAKGVEAFEK YVAEESTNAN WKPEEAKKFI EDVKAKAK |
-Macromolecule #3: ATPTB3
| Macromolecule | Name: ATPTB3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Trypanosoma brucei brucei (eukaryote) Trypanosoma brucei brucei (eukaryote) |
| Molecular weight | Theoretical: 27.646643 KDa |
| Sequence | String: MSKQLTFISA GATAAVLQSA SAIVSKVAGG RVQTKTAKEA GRHAVVVGPE TPIGVHTAVT EVPKSAQDPL FSGVSTVVVR AVLPRAAPD SVQLRDALDV YASAGIDTKE EVRSATEAFK KSAEVAVGKA KAKGVKRIVL VVKQASKHNC INELFKKIST E TIESAGLT ...String: MSKQLTFISA GATAAVLQSA SAIVSKVAGG RVQTKTAKEA GRHAVVVGPE TPIGVHTAVT EVPKSAQDPL FSGVSTVVVR AVLPRAAPD SVQLRDALDV YASAGIDTKE EVRSATEAFK KSAEVAVGKA KAKGVKRIVL VVKQASKHNC INELFKKIST E TIESAGLT TEVVGTAAVA NQLIVNPESL GVVLLNDVAA TEQIELAFAG VVGGVSRVYH TVEGGKISAG HSFKSVALAV AQ ELRELGL SSEADKVEAA ASKNPRAVVS AL |
-Macromolecule #4: subunit-8
| Macromolecule | Name: subunit-8 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Trypanosoma brucei brucei (eukaryote) Trypanosoma brucei brucei (eukaryote) |
| Molecular weight | Theoretical: 13.750958 KDa |
| Sequence | String: MLRRLGANVS NMARPMNKYA VTVSPRRHLE PMSTWYLASW AMVWYYAFFF WMPMVWTDIM VPSFVYNKLP VIHFLQEKRA EQKLRRVLD ETYTEWTEEL DQAHVTDAIT RSLNI |
-Macromolecule #5: subunit-d
| Macromolecule | Name: subunit-d / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Trypanosoma brucei brucei (eukaryote) Trypanosoma brucei brucei (eukaryote) |
| Molecular weight | Theoretical: 43.379324 KDa |
| Sequence | String: MRRVSSPNIT IQSVRWISGV SPLLYFPPTT TSTTNREDQI NKNTNIAIQM IKRYKGEVPP HYTRKSSATI EQVEKEIDAL LGGAEKLRK TSTDDQPMDK LTLMERCLRH ALWSYHKEEG RYDFDQIGRW VVYTPEDEVK LAQLKREVEA KEKLAALRKR R EEEGLPGG ...String: MRRVSSPNIT IQSVRWISGV SPLLYFPPTT TSTTNREDQI NKNTNIAIQM IKRYKGEVPP HYTRKSSATI EQVEKEIDAL LGGAEKLRK TSTDDQPMDK LTLMERCLRH ALWSYHKEEG RYDFDQIGRW VVYTPEDEVK LAQLKREVEA KEKLAALRKR R EEEGLPGG PVPRINWPQE YSSFIDREPV VAKRIRYDTL ASTTLERDEK QIESTLQQYR RASQDKRLDD LVDLLERFKP VL AREAIMQ RLTIKHLEGQ LGVWRYMDWC PEVRDRAELE VDITGWQWWS PLEERRLLPV RLRSVNEVRE IMSKTQAKKS AEA AERNPI VTQTSTGDNA RDRLLKEVLA LQARINQRDE VEPSQTEQKK KAHH |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.6 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 33.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z
Z Y
Y X
X