[English] 日本語
 Yorodumi
Yorodumi- EMDB-14922: cryo-EM structure of omicron spike in complex with de novo design... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | ||||||||||||||||||
 Map data Map data | vhf | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / entry receptor-mediated virion attachment to host cell / receptor-mediated endocytosis of virus by host cell / Attachment and Entry /  membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell /  receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||||||||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus 2 / Severe acute respiratory syndrome coronavirus 2 /   Drosophila melanogaster (fruit fly) / Drosophila melanogaster (fruit fly) /   Tequatrovirus T4 Tequatrovirus T4 | ||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.8 Å cryo EM / Resolution: 2.8 Å | ||||||||||||||||||
 Authors Authors | Pablo G / Sarah W / Alexandra VH / Anthony M / Andreas S / Zander H / Dongchun N / Shuguang T / Freyr S / Casper G ...Pablo G / Sarah W / Alexandra VH / Anthony M / Andreas S / Zander H / Dongchun N / Shuguang T / Freyr S / Casper G / Priscilla T / Alexandra T / Stephane R / Sandrine G / Jane M / Aaron P / Zepeng X / Yan C / Pu H / George G / Elisa O / Beat F / Didier T / Henning S / Michael B / Bruno EC | ||||||||||||||||||
| Funding support | European Union,  Switzerland, 5 items Switzerland, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: De novo design of protein interactions with learned surface fingerprints. Authors: Pablo Gainza / Sarah Wehrle / Alexandra Van Hall-Beauvais / Anthony Marchand / Andreas Scheck / Zander Harteveld / Stephen Buckley / Dongchun Ni / Shuguang Tan / Freyr Sverrisson / Casper ...Authors: Pablo Gainza / Sarah Wehrle / Alexandra Van Hall-Beauvais / Anthony Marchand / Andreas Scheck / Zander Harteveld / Stephen Buckley / Dongchun Ni / Shuguang Tan / Freyr Sverrisson / Casper Goverde / Priscilla Turelli / Charlène Raclot / Alexandra Teslenko / Martin Pacesa / Stéphane Rosset / Sandrine Georgeon / Jane Marsden / Aaron Petruzzella / Kefang Liu / Zepeng Xu / Yan Chai / Pu Han / George F Gao / Elisa Oricchio / Beat Fierz / Didier Trono / Henning Stahlberg / Michael Bronstein / Bruno E Correia /    Abstract: Physical interactions between proteins are essential for most biological processes governing life. However, the molecular determinants of such interactions have been challenging to understand, even ...Physical interactions between proteins are essential for most biological processes governing life. However, the molecular determinants of such interactions have been challenging to understand, even as genomic, proteomic and structural data increase. This knowledge gap has been a major obstacle for the comprehensive understanding of cellular protein-protein interaction networks and for the de novo design of protein binders that are crucial for synthetic biology and translational applications. Here we use a geometric deep-learning framework operating on protein surfaces that generates fingerprints to describe geometric and chemical features that are critical to drive protein-protein interactions. We hypothesized that these fingerprints capture the key aspects of molecular recognition that represent a new paradigm in the computational design of novel protein interactions. As a proof of principle, we computationally designed several de novo protein binders to engage four protein targets: SARS-CoV-2 spike, PD-1, PD-L1 and CTLA-4. Several designs were experimentally optimized, whereas others were generated purely in silico, reaching nanomolar affinity with structural and mutational characterization showing highly accurate predictions. Overall, our surface-centric approach captures the physical and chemical determinants of molecular recognition, enabling an approach for the de novo design of protein interactions and, more broadly, of artificial proteins with function. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14922.map.gz emd_14922.map.gz | 79.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14922-v30.xml emd-14922-v30.xml emd-14922.xml emd-14922.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
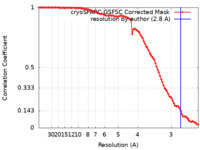
| FSC (resolution estimation) |  emd_14922_fsc.xml emd_14922_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14922.png emd_14922.png | 40.9 KB | ||
| Others |  emd_14922_half_map_1.map.gz emd_14922_half_map_1.map.gz emd_14922_half_map_2.map.gz emd_14922_half_map_2.map.gz | 77.8 MB 77.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14922 http://ftp.pdbj.org/pub/emdb/structures/EMD-14922 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14922 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14922 | HTTPS FTP |
-Related structure data
| Related structure data |  7zrvMC  7xadC  7xyqC  7zsdC  7zssC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14922.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14922.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | vhf | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2342 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: gfgf
| File | emd_14922_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | gfgf | ||||||||||||
| Projections & Slices |
| ||||||||||||
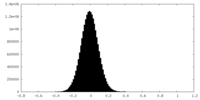
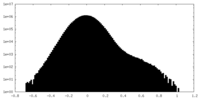
| Density Histograms |
-Half map: fgfg
| File | emd_14922_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | fgfg | ||||||||||||
| Projections & Slices |
| ||||||||||||
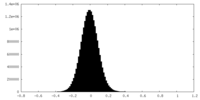
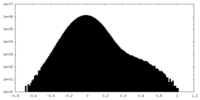
| Density Histograms |
- Sample components
Sample components
-Entire : omicron spike in complex with de novo designed binder, full map
| Entire | Name: omicron spike in complex with de novo designed binder, full map |
|---|---|
| Components |
|
-Supramolecule #1: omicron spike in complex with de novo designed binder, full map
| Supramolecule | Name: omicron spike in complex with de novo designed binder, full map type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: omicron spike
| Supramolecule | Name: omicron spike / type: complex / ID: 2 / Chimera: Yes / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
-Supramolecule #3: de novo designed binder
| Supramolecule | Name: de novo designed binder / type: complex / ID: 3 / Chimera: Yes / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
-Macromolecule #1: Spike glycoprotein,Envelope glycoprotein
| Macromolecule | Name: Spike glycoprotein,Envelope glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tequatrovirus T4 Tequatrovirus T4 |
| Molecular weight | Theoretical: 142.558094 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ PFLMDLEGKQ GNFKNLREFV FKNIDGYFKI YSKHTPIIVR EPEDLPQGFS ALEPLVDLPI GINITRFQTL LA LHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPT ESIVRF PNITNLCPFD EVFNATRFAS VYAWNRKRIS NCVADYSVLY NLAPFFTFKC YGVSPTKLND LCFTNVYADS FVIR GDEVR QIAPGQTGNI ADYNYKLPDD FTGCVIAWNS NKLDSKVSGN YNYLYRLFRK SNLKPFERDI STEIYQAGNK PCNGV AGFN CYFPLRSYSF RPTYGVGHQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLK GTGVLTESNK KFLPFQ QFG RDIADTTDAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQGVN CTEVPVAIHA DQLTPTWRVY STGSNVF QT RAGCLIGAEY VNNSYECDIP IGAGICASYQ TQTKSHGSAS SVASQSIIAY TMSLGAENSV AYSNNSIAIP TNFTISVT T EILPVSMTKT SVDCTMYICG DSTECSNLLL QYGSFCTQLK RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KYFGGFNFS QILPDPSKPS KRSFIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFKGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGA ALQIPFAMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTA SALGKLQDVV NHNAQALNTL V KQLSSKFG AISSVLNDIF SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FC GKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVS GNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQEL GKYEQ GSGYIPEAPR DGQAYVRKDG EWVLLSTFLG RSLEVLFQGP GHHHHHHHHS AWSHPQFEKG GGSGGGGSGG SAWSH PQFE K |
-Macromolecule #2: de novo designed binder
| Macromolecule | Name: de novo designed binder / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
| Molecular weight | Theoretical: 8.829904 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: ETGASSTNML EALQQRLQFY HGQVARAALE NNSGKARRFG RIVKQYEDAI KLYKAGKPVP YDELPVPPGF GGSENLYFQ |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 16 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Specialist optics | Energy filter - Name: TFS Selectris X |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average exposure time: 5.82 sec. / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 33.8 |
| Output model |  PDB-7zrv: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)