[English] 日本語
 Yorodumi
Yorodumi- EMDB-14697: M. tuberculosis RNA polymerase in complex with sigma-b initiation... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | M. tuberculosis RNA polymerase in complex with sigma-b initiation factor: octameric assembly of (alpha)2-beta-beta'-omega-sigB protomers. | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  RNA polymerase / RNA polymerase /  self-assembly / self-assembly /  octamer / octamer /  tuberculosis / tuberculosis /  stress response / stress response /  hibernation / hibernation /  TRANSCRIPTION TRANSCRIPTION | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationAntimicrobial action and antimicrobial resistance in Mtb /  sigma factor activity / peptidoglycan-based cell wall / sigma factor activity / peptidoglycan-based cell wall /  DNA-directed RNA polymerase complex / DNA-templated transcription initiation / DNA-directed RNA polymerase complex / DNA-templated transcription initiation /  ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity / ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity /  DNA-directed RNA polymerase / DNA-directed RNA polymerase /  protein dimerization activity / response to antibiotic ...Antimicrobial action and antimicrobial resistance in Mtb / protein dimerization activity / response to antibiotic ...Antimicrobial action and antimicrobial resistance in Mtb /  sigma factor activity / peptidoglycan-based cell wall / sigma factor activity / peptidoglycan-based cell wall /  DNA-directed RNA polymerase complex / DNA-templated transcription initiation / DNA-directed RNA polymerase complex / DNA-templated transcription initiation /  ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity / ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity /  DNA-directed RNA polymerase / DNA-directed RNA polymerase /  protein dimerization activity / response to antibiotic / DNA-templated transcription / magnesium ion binding / protein dimerization activity / response to antibiotic / DNA-templated transcription / magnesium ion binding /  DNA binding / zinc ion binding / DNA binding / zinc ion binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||||||||
| Biological species |   Mycobacterium tuberculosis (bacteria) Mycobacterium tuberculosis (bacteria) | ||||||||||||
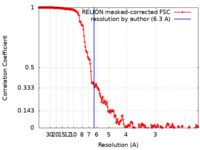
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 6.3 Å cryo EM / Resolution: 6.3 Å | ||||||||||||
 Authors Authors | Bron P / Trapani S / Lai Kee Him J / Brodolin K / Morichaud Z / Vishwakarma R | ||||||||||||
| Funding support |  France, 3 items France, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization. Authors: Zakia Morichaud / Stefano Trapani / Rishi K Vishwakarma / Laurent Chaloin / Corinne Lionne / Joséphine Lai-Kee-Him / Patrick Bron / Konstantin Brodolin /   Abstract: Self-assembly of macromolecules into higher-order symmetric structures is fundamental for the regulation of biological processes. Higher-order symmetric structure self-assembly by the gene expression ...Self-assembly of macromolecules into higher-order symmetric structures is fundamental for the regulation of biological processes. Higher-order symmetric structure self-assembly by the gene expression machinery, such as bacterial DNA-dependent RNA polymerase (RNAP), has never been reported before. Here, we show that the stress-response σ factor from the human pathogen, Mycobacterium tuberculosis, induces the RNAP holoenzyme oligomerization into a supramolecular complex composed of eight RNAP units. Cryo-electron microscopy revealed a pseudo-symmetric structure of the RNAP octamer in which RNAP protomers are captured in an auto-inhibited state and display an open-clamp conformation. The structure shows that σ is sequestered by the RNAP flap and clamp domains. The transcriptional activator RbpA prevented octamer formation by promoting the initiation-competent RNAP conformation. Our results reveal that a non-conserved region of σ is an allosteric controller of transcription initiation and demonstrate how basal transcription factors can regulate gene expression by modulating the RNAP holoenzyme assembly and hibernation. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization Authors: Morichaud Z / Trapani S / Vishwakarma R / Chaloin L / Lionne C / Lai-Kee-Him J / Bron P / Brodolin K | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14697.map.gz emd_14697.map.gz | 398.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14697-v30.xml emd-14697-v30.xml emd-14697.xml emd-14697.xml | 25.4 KB 25.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14697_fsc.xml emd_14697_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14697.png emd_14697.png | 189.4 KB | ||
| Filedesc metadata |  emd-14697.cif.gz emd-14697.cif.gz | 7.6 KB | ||
| Others |  emd_14697_half_map_1.map.gz emd_14697_half_map_1.map.gz emd_14697_half_map_2.map.gz emd_14697_half_map_2.map.gz | 401.4 MB 402.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14697 http://ftp.pdbj.org/pub/emdb/structures/EMD-14697 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14697 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14697 | HTTPS FTP |
-Related structure data
| Related structure data |  7zf2C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14697.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14697.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
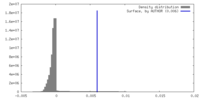
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_14697_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
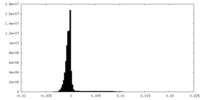
| Density Histograms |
-Half map: #2
| File | emd_14697_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
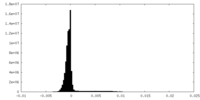
| Density Histograms |
- Sample components
Sample components
-Entire : M. tuberculosis RNA polymerase in complex with sigma-b initiation...
| Entire | Name: M. tuberculosis RNA polymerase in complex with sigma-b initiation factor: octameric assembly of (alpha)2-beta-beta'-omega-sigB protomers. |
|---|---|
| Components |
|
-Supramolecule #1: M. tuberculosis RNA polymerase in complex with sigma-b initiation...
| Supramolecule | Name: M. tuberculosis RNA polymerase in complex with sigma-b initiation factor: octameric assembly of (alpha)2-beta-beta'-omega-sigB protomers. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 402 KDa |
-Macromolecule #1: DNA-directed RNA polymerase subunit alpha
| Macromolecule | Name: DNA-directed RNA polymerase subunit alpha / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number:  DNA-directed RNA polymerase DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis (bacteria) Mycobacterium tuberculosis (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MLISQRPTLS EDVLTDNRSQ FVIEPLEPGF GYTLGNSLRR TLLSSIPGAA VTSIRIDGVL HEFTTVPGVK EDVTEIILNL KSLVVSSEED EPVTMYLRKQ GPGEVTAGDI VPPAGVTVHN PGMHIATLND KGKLEVELVV ERGRGYVPAV QNRASGAEIG RIPVDSIYSP ...String: MLISQRPTLS EDVLTDNRSQ FVIEPLEPGF GYTLGNSLRR TLLSSIPGAA VTSIRIDGVL HEFTTVPGVK EDVTEIILNL KSLVVSSEED EPVTMYLRKQ GPGEVTAGDI VPPAGVTVHN PGMHIATLND KGKLEVELVV ERGRGYVPAV QNRASGAEIG RIPVDSIYSP VLKVTYKVDA TRVEQRTDFD KLILDVETKN SISPRDALAS AGKTLVELFG LARELNVEAE GIEIGPSPAE ADHIASFALP IDDLDLTVRS YNCLKREGVH TVGELVARTE SDLLDIRNFG QKSIDEVKIK LHQLGLSLKD SPPSFDPSEV AGYDVATGTW STEGAYDEQD YAETEQL UniProtKB:  DNA-directed RNA polymerase subunit alpha DNA-directed RNA polymerase subunit alpha |
-Macromolecule #2: DNA-directed RNA polymerase subunit beta
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO / EC number:  DNA-directed RNA polymerase DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis (bacteria) Mycobacterium tuberculosis (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MVLADSRQSK TAASPSPSRP QSSSNNSVPG APNRVSFAKL REPLEVPGLL DVQTDSFEWL IGSPRWRESA AERGDVNPVG GLEEVLYELS PIEDFSGSMS LSFSDPRFDD VKAPVDECKD KDMTYAAPLF VTAEFINNNT GEIKSQTVFM GDFPMMTEKG TFIINGTERV ...String: MVLADSRQSK TAASPSPSRP QSSSNNSVPG APNRVSFAKL REPLEVPGLL DVQTDSFEWL IGSPRWRESA AERGDVNPVG GLEEVLYELS PIEDFSGSMS LSFSDPRFDD VKAPVDECKD KDMTYAAPLF VTAEFINNNT GEIKSQTVFM GDFPMMTEKG TFIINGTERV VVSQLVRSPG VYFDETIDKS TDKTLHSVKV IPSRGAWLEF DVDKRDTVGV RIDRKRRQPV TVLLKALGWT SEQIVERFGF SEIMRSTLEK DNTVGTDEAL LDIYRKLRPG EPPTKESAQT LLENLFFKEK RYDLARVGRY KVNKKLGLHV GEPITSSTLT EEDVVATIEY LVRLHEGQTT MTVPGGVEVP VETDDIDHFG NRRLRTVGEL IQNQIRVGMS RMERVVRERM TTQDVEAITP QTLINIRPVV AAIKEFFGTS QLSQFMDQNN PLSGLTHKRR LSALGPGGLS RERAGLEVRD VHPSHYGRMC PIETPEGPNI GLIGSLSVYA RVNPFGFIET PYRKVVDGVV SDEIVYLTAD EEDRHVVAQA NSPIDADGRF VEPRVLVRRK AGEVEYVPSS EVDYMDVSPR QMVSVATAMI PFLEHDDANR ALMGANMQRQ AVPLVRSEAP LVGTGMELRA AIDAGDVVVA EESGVIEEVS ADYITVMHDN GTRRTYRMRK FARSNHGTCA NQCPIVDAGD RVEAGQVIAD GPCTDDGEMA LGKNLLVAIM PWEGHNYEDA IILSNRLVEE DVLTSIHIEE HEIDARDTKL GAEEITRDIP NISDEVLADL DERGIVRIGA EVRDGDILVG KVTPKGETEL TPEERLLRAI FGEKAREVRD TSLKVPHGES GKVIGIRVFS REDEDELPAG VNELVRVYVA QKRKISDGDK LAGRHGNKGV IGKILPVEDM PFLADGTPVD IILNTHGVPR RMNIGQILET HLGWCAHSGW KVDAAKGVPD WAARLPDELL EAQPNAIVST PVFDGAQEAE LQGLLSCTLP NRDGDVLVDA DGKAMLFDGR SGEPFPYPVT VGYMYIMKLH HLVDDKIHAR STGPYSMITQ QPLGGKAQFG GQRFGEMECW AMQAYGAAYT LQELLTIKSD DTVGRVKVYE AIVKGENIPE PGIPESFKVL LKELQSLCLN VEVLSSDGAA IELREGEDED LERAAANLGI NLSRNESASV EDLA UniProtKB:  DNA-directed RNA polymerase subunit beta DNA-directed RNA polymerase subunit beta |
-Macromolecule #3: DNA-directed RNA polymerase subunit beta'
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta' / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO / EC number:  DNA-directed RNA polymerase DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis (bacteria) Mycobacterium tuberculosis (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: VNFFDELRIG LATAEDIRQW SYGEVKKPET INYRTLKPEK DGLFCEKIFG PTRDWECYCG KYKRVRFKGI ICERCGVEVT RAKVRRERMG HIELAAPVTH IWYFKGVPSR LGYLLDLAPK DLEKIIYFAA YVITSVDEEM RHNELSTLEA EMAVERKAVE DQRDGELEAR ...String: VNFFDELRIG LATAEDIRQW SYGEVKKPET INYRTLKPEK DGLFCEKIFG PTRDWECYCG KYKRVRFKGI ICERCGVEVT RAKVRRERMG HIELAAPVTH IWYFKGVPSR LGYLLDLAPK DLEKIIYFAA YVITSVDEEM RHNELSTLEA EMAVERKAVE DQRDGELEAR AQKLEADLAE LEAEGAKADA RRKVRDGGER EMRQIRDRAQ RELDRLEDIW STFTKLAPKQ LIVDENLYRE LVDRYGEYFT GAMGAESIQK LIENFDIDAE AESLRDVIRN GKGQKKLRAL KRLKVVAAFQ QSGNSPMGMV LDAVPVIPPE LRPMVQLDGG RFATSDLNDL YRRVINRNNR LKRLIDLGAP EIIVNNEKRM LQESVDALFD NGRRGRPVTG PGNRPLKSLS DLLKGKQGRF RQNLLGKRVD YSGRSVIVVG PQLKLHQCGL PKLMALELFK PFVMKRLVDL NHAQNIKSAK RMVERQRPQV WDVLEEVIAE HPVLLNRAPT LHRLGIQAFE PMLVEGKAIQ LHPLVCEAFN ADFDGDQMAV HLPLSAEAQA EARILMLSSN NILSPASGRP LAMPRLDMVT GLYYLTTEVP GDTGEYQPAS GDHPETGVYS SPAEAIMAAD RGVLSVRAKI KVRLTQLRPP VEIEAELFGH SGWQPGDAWM AETTLGRVMF NELLPLGYPF VNKQMHKKVQ AAIINDLAER YPMIVVAQTV DKLKDAGFYW ATRSGVTVSM ADVLVPPRKK EILDHYEERA DKVEKQFQRG ALNHDERNEA LVEIWKEATD EVGQALREHY PDDNPIITIV DSGATGNFTQ TRTLAGMKGL VTNPKGEFIP RPVKSSFREG LTVLEYFINT HGARKGLADT ALRTADSGYL TRRLVDVSQD VIVREHDCQT ERGIVVELAE RAPDGTLIRD PYIETSAYAR TLGTDAVDEA GNVIVERGQD LGDPEIDALL AAGITQVKVR SVLTCATSTG VCATCYGRSM ATGKLVDIGE AVGIVAAQSI GEPGTQLTMR TFHQGGVGED ITGGLPRVQE LFEARVPRGK APIADVTGRV RLEDGERFYK ITIVPDDGGE EVVYDKISKR QRLRVFKHED GSERVLSDGD HVEVGQQLME GSADPHEVLR VQGPREVQIH LVREVQEVYR AQGVSIHDKH IEVIVRQMLR RVTIIDSGST EFLPGSLIDR AEFEAENRRV VAEGGEPAAG RPVLMGITKA SLATDSWLSA ASFQETTRVL TDAAINCRSD KLNGLKENVI IGKLIPAGTG INRYRNIAVQ PTEEARAAAY TIPSYEDQYY SPDFGAATGA AVPLDDYGYS DYRHHHHHH UniProtKB:  DNA-directed RNA polymerase subunit beta' DNA-directed RNA polymerase subunit beta' |
-Macromolecule #4: DNA-directed RNA polymerase subunit omega
| Macromolecule | Name: DNA-directed RNA polymerase subunit omega / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO / EC number:  DNA-directed RNA polymerase DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis (bacteria) Mycobacterium tuberculosis (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSISQSDASL AAVPAVDQFD PSSGASGGYD TPLGITNPPI DELLDRVSSK YALVIYAAKR ARQINDYYNQ LGEGILEYVG PLVEPGLQEK PLSIALREIH ADLLEHTEGE UniProtKB:  DNA-directed RNA polymerase subunit omega DNA-directed RNA polymerase subunit omega |
-Macromolecule #5: RNA polymerase sigma factor SigB
| Macromolecule | Name: RNA polymerase sigma factor SigB / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis (bacteria) Mycobacterium tuberculosis (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MADAPTRATT SRVDSDLDAQ SPAADLVRVY LNGIGKTALL NAAGEVELAK RIEAGLYAEH LLETRKRLGE NRKRDLAAVV RDGEAARRHL LEANLRLVVS LAKRYTGRGM PLLDLIQEGN LGLIRAMEKF DYTKGFKFST YATWWIRQAI ...String: MGSSHHHHHH SSGLVPRGSH MADAPTRATT SRVDSDLDAQ SPAADLVRVY LNGIGKTALL NAAGEVELAK RIEAGLYAEH LLETRKRLGE NRKRDLAAVV RDGEAARRHL LEANLRLVVS LAKRYTGRGM PLLDLIQEGN LGLIRAMEKF DYTKGFKFST YATWWIRQAI TRGMADQSRT IRLPVHLVEQ VNKLARIKRE MHQHLGREAT DEELAAESGI PIDKINDLLE HSRDPVSLDM PVGSEEEAPL GDFIEDAEAM SAENAVIAEL LHTDIRSVLA TLDEREHQVI RLRFGLDDGQ PRTLDQIGKL FGLSRERVRQ IERDVMSKLR HGERADRLRS YAS UniProtKB: RNA polymerase sigma factor SigB |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.21 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 7.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 Bright-field microscopy / Nominal defocus max: 7.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 3064 / Average exposure time: 8.0 sec. / Average electron dose: 49.6 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)