+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | gp6/gp15/gp16 connector complex of bacteriophage SPP1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationviral DNA genome packaging, headful / symbiont genome ejection through host cell envelope, long flexible tail mechanism / viral portal complex / viral procapsid /  virion component virion componentSimilarity search - Function | |||||||||
| Biological species |   Bacillus subtilis (bacteria) Bacillus subtilis (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.7 Å cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Orlov I / Roche S / Tavares P / Orlova EV | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: CryoEM structure and assembly mechanism of a bacterial virus genome gatekeeper. Authors: Igor Orlov / Stéphane Roche / Sandrine Brasilès / Natalya Lukoyanova / Marie-Christine Vaney / Paulo Tavares / Elena V Orlova /   Abstract: Numerous viruses package their dsDNA genome into preformed capsids through a portal gatekeeper that is subsequently closed. We report the structure of the DNA gatekeeper complex of bacteriophage SPP1 ...Numerous viruses package their dsDNA genome into preformed capsids through a portal gatekeeper that is subsequently closed. We report the structure of the DNA gatekeeper complex of bacteriophage SPP1 (gp6gp15gp16) in the post-DNA packaging state at 2.7 Å resolution obtained by single particle cryo-electron microscopy. Comparison of the native SPP1 complex with assembly-naïve structures of individual components uncovered the complex program of conformational changes leading to its assembly. After DNA packaging, gp15 binds via its C-terminus to the gp6 oligomer positioning gp15 subunits for oligomerization. Gp15 refolds its inner loops creating an intersubunit β-barrel that establishes different types of contacts with six gp16 subunits. Gp16 binding and oligomerization is accompanied by folding of helices that close the portal channel to keep the viral genome inside the capsid. This mechanism of assembly has broad functional and evolutionary implications for viruses of the prokaryotic tailed viruses-herpesviruses lineage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14509.map.gz emd_14509.map.gz | 351 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14509-v30.xml emd-14509-v30.xml emd-14509.xml emd-14509.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14509.png emd_14509.png | 170 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14509 http://ftp.pdbj.org/pub/emdb/structures/EMD-14509 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14509 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14509 | HTTPS FTP |
-Related structure data
| Related structure data |  7z4wMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14509.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14509.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.69 Å | ||||||||||||||||||||||||||||||||||||
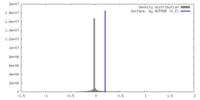
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : gp6(12) gp15(12) gp16(6) portal complex (connector) of SPP1 bacte...
| Entire | Name: gp6(12) gp15(12) gp16(6) portal complex (connector) of SPP1 bacteriophage |
|---|---|
| Components |
|
-Supramolecule #1: gp6(12) gp15(12) gp16(6) portal complex (connector) of SPP1 bacte...
| Supramolecule | Name: gp6(12) gp15(12) gp16(6) portal complex (connector) of SPP1 bacteriophage type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Bacillus subtilis (bacteria) / Strain: YB886 Bacillus subtilis (bacteria) / Strain: YB886 |
| Molecular weight | Theoretical: 0.902 kDa/nm |
-Macromolecule #1: Portal protein
| Macromolecule | Name: Portal protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bacillus subtilis (bacteria) Bacillus subtilis (bacteria) |
| Molecular weight | Theoretical: 57.390277 KDa |
| Sequence | String: MADIYPLGKT HTEELNEIIV ESAKEIAEPD TTMIQKLIDE HNPEPLLKGV RYYMCENDIE KKRRTYYDAA GQQLVDDTKT NNRTSHAWH KLFVDQKTQY LVGEPVTFTS DNKTLLEYVN ELADDDFDDI LNETVKNMSN KGIEYWHPFV DEEGEFDYVI F PAEEMIVV ...String: MADIYPLGKT HTEELNEIIV ESAKEIAEPD TTMIQKLIDE HNPEPLLKGV RYYMCENDIE KKRRTYYDAA GQQLVDDTKT NNRTSHAWH KLFVDQKTQY LVGEPVTFTS DNKTLLEYVN ELADDDFDDI LNETVKNMSN KGIEYWHPFV DEEGEFDYVI F PAEEMIVV YKDNTRRDIL FALRYYSYKG IMGEETQKAE LYTDTHVYYY EKIDGVYQMD YSYGENNPRP HMTKGGQAIG WG RVPIIPF KNNEEMVSDL KFYKDLIDNY DSITSSTMDS FSDFQQIVYV LKNYDGENPK EFTANLRYHS VIKVSGDGGV DTL RAEIPV DSAAKELERI QDELYKSAQA VDNSPETIGG GATGPALENL YALLDLKANM AERKIRAGLR LFFWFFAEYL RNTG KGDFN PDKELTMTFT RTRIQNDSEI VQSLVQGVTG GIMSKETAVA RNPFVQDPEE ELARIEEEMN QYAEMQGNLL DDEGG DDDL EEDDPNAGAA ESGGAGQVS |
-Macromolecule #2: Head completion protein gp15
| Macromolecule | Name: Head completion protein gp15 / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bacillus subtilis (bacteria) Bacillus subtilis (bacteria) |
| Molecular weight | Theoretical: 11.629402 KDa |
| Sequence | String: MDIQRVKRLL SITNDKHDEY LTEMVPLLVE FAKDECHNPF IDKDGNESIP SGVLIFVAKA AQFYMTNAGL TGRSMDTVSY NFATEIPST ILKKLNPYRK MAR |
-Macromolecule #3: Head completion protein gp16
| Macromolecule | Name: Head completion protein gp16 / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bacillus subtilis (bacteria) Bacillus subtilis (bacteria) |
| Molecular weight | Theoretical: 12.55499 KDa |
| Sequence | String: MYEEFPDVIT FQSYVEQSNG EGGKTYKWVD EFTAAAHVQP ISQEEYYKAQ QLQTPIGYNI YTPYDDRIDK KMRVIYRGKI VTFIGDPVD LSGLQEITRI KGKEDGAYVG |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 406 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.64 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 96 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 120000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 3876 / Average exposure time: 1.0 sec. / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 520000 |
|---|---|
| Startup model | Type of model: EMDB MAP EMDB ID: Details: The map has been low pass filtered to 20 Angstrom. |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 2.0) |
| Final 3D classification | Number classes: 5 / Software - Name: cryoSPARC (ver. 2) Details: After three rounds of 2D classification in CryoSPARC 401,807 particle images were selected for the next two rounds of refinement. |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 2.0) |
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: cryoSPARC (ver. 2) ) / Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: cryoSPARC (ver. 2)Details: Local resolution variations in the reconstruction was estimated using ResMap Number images used: 401807 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Residue range: 254-344 |
|---|---|
| Details | Manual inspection of the residues in the complete gp6, gp15 and gp16 polypeptide chains tracing was done using COOT and refined in Phenix |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 90 |
| Output model |  PDB-7z4w: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)