[English] 日本語
 Yorodumi
Yorodumi- EMDB-14090: Unique vertex of the phicrAss001 virion with C5 symmetry imposed -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Unique vertex of the phicrAss001 virion with C5 symmetry imposed | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information viral capsid, decoration / viral capsid, decoration /  virion component / virion component /  viral capsid / symbiont entry into host cell / virion attachment to host cell viral capsid / symbiont entry into host cell / virion attachment to host cellSimilarity search - Function | |||||||||
| Biological species |  Bacteroides phage crAss001 (virus) Bacteroides phage crAss001 (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.32 Å cryo EM / Resolution: 3.32 Å | |||||||||
 Authors Authors | Bayfield OW / Shkoporov AN / Yutin N / Khokhlova EV / Smith JLR / Hawkins DEDP / Koonin EV / Hill C / Antson AA | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structural atlas of a human gut crassvirus. Authors: Oliver W Bayfield / Andrey N Shkoporov / Natalya Yutin / Ekaterina V Khokhlova / Jake L R Smith / Dorothy E D P Hawkins / Eugene V Koonin / Colin Hill / Alfred A Antson /    Abstract: CrAssphage and related viruses of the order Crassvirales (hereafter referred to as crassviruses) were originally discovered by cross-assembly of metagenomic sequences. They are the most abundant ...CrAssphage and related viruses of the order Crassvirales (hereafter referred to as crassviruses) were originally discovered by cross-assembly of metagenomic sequences. They are the most abundant viruses in the human gut, are found in the majority of individual gut viromes, and account for up to 95% of the viral sequences in some individuals. Crassviruses are likely to have major roles in shaping the composition and functionality of the human microbiome, but the structures and roles of most of the virally encoded proteins are unknown, with only generic predictions resulting from bioinformatic analyses. Here we present a cryo-electron microscopy reconstruction of Bacteroides intestinalis virus ΦcrAss001, providing the structural basis for the functional assignment of most of its virion proteins. The muzzle protein forms an assembly about 1 MDa in size at the end of the tail and exhibits a previously unknown fold that we designate the 'crass fold', that is likely to serve as a gatekeeper that controls the ejection of cargos. In addition to packing the approximately 103 kb of virus DNA, the ΦcrAss001 virion has extensive storage space for virally encoded cargo proteins in the capsid and, unusually, within the tail. One of the cargo proteins is present in both the capsid and the tail, suggesting a general mechanism for protein ejection, which involves partial unfolding of proteins during their extrusion through the tail. These findings provide a structural basis for understanding the mechanisms of assembly and infection of these highly abundant crassviruses. #1:  Journal: Res Sq / Year: 2023 Journal: Res Sq / Year: 2023Title: Structural atlas of the most abundant human gut virus Authors: Antson A / Bayfield O / Shkoporov A / Yutin N / Khokhlova E / Smith J / Hawkins D / Koonin E / Hill C | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14090.map.gz emd_14090.map.gz | 52.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14090-v30.xml emd-14090-v30.xml emd-14090.xml emd-14090.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14090_fsc.xml emd_14090_fsc.xml | 10.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14090.png emd_14090.png | 154.4 KB | ||
| Others |  emd_14090_additional_1.map.gz emd_14090_additional_1.map.gz emd_14090_additional_2.map.gz emd_14090_additional_2.map.gz | 29.8 MB 82.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14090 http://ftp.pdbj.org/pub/emdb/structures/EMD-14090 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14090 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14090 | HTTPS FTP |
-Related structure data
| Related structure data |  7qohMC  7qofC  7qogC  7qoiC  7qojC  7qokC  7qolC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14090.map.gz / Format: CCP4 / Size: 89.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14090.map.gz / Format: CCP4 / Size: 89.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3941 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_14090_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
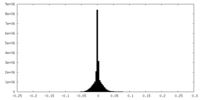
| Density Histograms |
-Additional map: #2
| File | emd_14090_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
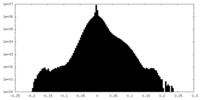
| Density Histograms |
- Sample components
Sample components
-Entire : Bacteroides phage crAss001
| Entire | Name:  Bacteroides phage crAss001 (virus) Bacteroides phage crAss001 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Bacteroides phage crAss001
| Supramolecule | Name: Bacteroides phage crAss001 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 / NCBI-ID: 2301731 / Sci species name: Bacteroides phage crAss001 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Major capsid protein gp32
| Macromolecule | Name: Major capsid protein gp32 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides phage crAss001 (virus) Bacteroides phage crAss001 (virus) |
| Molecular weight | Theoretical: 57.098598 KDa |
| Sequence | String: MAGKLGKFQM LGFQHWKGLT SDNHLGAIFQ QAPQKATNLM VQLLAFYRGK SLDTFLNSFP TREFEDDNEY YWDVIGSSRR NIPLVEARD ENGVVVAANA ANVGVGTSPF YLVFPEDWFA DGEVIVGNLN QVYPFRILGD ARMEGTNAVY KVELMGGNTQ G VPAERLQQ ...String: MAGKLGKFQM LGFQHWKGLT SDNHLGAIFQ QAPQKATNLM VQLLAFYRGK SLDTFLNSFP TREFEDDNEY YWDVIGSSRR NIPLVEARD ENGVVVAANA ANVGVGTSPF YLVFPEDWFA DGEVIVGNLN QVYPFRILGD ARMEGTNAVY KVELMGGNTQ G VPAERLQQ GERFSIEFAP VEKELSRKVG DVRFTSPVSM RNEWTTIRIQ HKVAGNKLNK KLAMGIPMVR NLESGKQVKD TA NMWMHYV DWEVELQFDE YKNNAMAWGT SNRNLNGEYM NFGKSGNAIK TGAGIFEQTE VANTMYYNTF SLKLLEDALY ELS ASKLAM DDRLFVIKTG ERGAIQFHKE VLKTVSGWTT FVLDNNSTRV VEKVQSRLHS NALSAGFQFV EYKAPNGVRV RLDV DPFYD DPVRNKILHP MGGVAFSYRY DIWYIGTMDQ PNIFKCKIKG DNEYRGYQWG IRNPFTGQKG NPYMSFDEDS AVIHR MATL GVCVLDPTRT MSLIPAILQG |
-Macromolecule #2: Auxiliary capsid protein gp36
| Macromolecule | Name: Auxiliary capsid protein gp36 / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides phage crAss001 (virus) Bacteroides phage crAss001 (virus) |
| Molecular weight | Theoretical: 36.154094 KDa |
| Sequence | String: MVISINQVRQ LYVAKALKAN TAALTTAGDI VPKADTAKTT LYFQSMSPAG IVASDKINLK HVLYAKATPS EALAHKLVRY SVTLDADVS ATPVAGQNYI LRLAFRQYIG LSEEDQYFKY GEVIARSGMT ASDFYKKMAI SLAKNLENKT ESTPLVNIYL I SAAAASTD ...String: MVISINQVRQ LYVAKALKAN TAALTTAGDI VPKADTAKTT LYFQSMSPAG IVASDKINLK HVLYAKATPS EALAHKLVRY SVTLDADVS ATPVAGQNYI LRLAFRQYIG LSEEDQYFKY GEVIARSGMT ASDFYKKMAI SLAKNLENKT ESTPLVNIYL I SAAAASTD VPVTSATKES DLTATDYNQI IIEETEQPWV LGMMPQAFIP FTPQFLTITV DGEDRLWGVA TVVTPTKTVP DG HLIADLE YFCMGARGDI YRGMGYPNII KTTYLVDPGA VYDVLDIHYF YTGSNESVQK SEKTITLVAV DDGSHTAMNA LIG AINTAS GLTIATL |
-Macromolecule #3: Portal vertex capsid protein gp57
| Macromolecule | Name: Portal vertex capsid protein gp57 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides phage crAss001 (virus) Bacteroides phage crAss001 (virus) |
| Molecular weight | Theoretical: 11.32708 KDa |
| Sequence | String: MAGQQGIYCA PDNIVPNRDR VDVGCAPDGA MQLWVMEYEV TGIGKGCAMC KAINPQQAEM LLKSNGIYNG SSYLYKVTRI EQVIVPPCN GLMAEQVVTY KDVVS |
-Macromolecule #4: Head fiber trimer protein gp21
| Macromolecule | Name: Head fiber trimer protein gp21 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides phage crAss001 (virus) Bacteroides phage crAss001 (virus) |
| Molecular weight | Theoretical: 10.294763 KDa |
| Sequence | String: MKTLRTLKIS PNAPDINSVW LYKGTMKYFN NGEWETIGGE SEPYVLPAAT TSTIGGVKKA TNVGNLATGA ELATVVTKVN AILSALKVA DIMVEDAN |
-Macromolecule #5: Portal protein gp20
| Macromolecule | Name: Portal protein gp20 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides phage crAss001 (virus) Bacteroides phage crAss001 (virus) |
| Molecular weight | Theoretical: 93.122211 KDa |
| Sequence | String: MADFLNFPRQ MLPFSKKTKQ WRKDCLLWAN QKTFFNYSLV RKSVIHKKIN YDLLNGRLHM SDLELVLNPD GIKAAYIPDR LQHYPIMNS KLNVLRGEES KRVFDFKVVV TNPNAISEIE DNKKNELLQR LQEMITDTSI SEDEYNIKLE KLNDYYTYEW Q DIREVRAN ...String: MADFLNFPRQ MLPFSKKTKQ WRKDCLLWAN QKTFFNYSLV RKSVIHKKIN YDLLNGRLHM SDLELVLNPD GIKAAYIPDR LQHYPIMNS KLNVLRGEES KRVFDFKVVV TNPNAISEIE DNKKNELLQR LQEMITDTSI SEDEYNIKLE KLNDYYTYEW Q DIREVRAN ELLNHYIKEY DIPLIFNNGF MDAMTCGEEI YQCDIVGGEP VIERVNPLKI RIFKSGYSNK VEDADMIILE DY WSPGRVI DTYYDVLSPK DIKYIETMPD YIGQGAVDQM DNIDERYGFV NQNMIGDEIT VRDGTYFFDP ANLFTEGIAN SLL PYDLAG NLRVLRLYWK SKRKILKVKS YDPETGEEEW NFYPENYVVN KEAGEEVQSF WVNEAWEGTM IGNEIFVNMR PRLI QYNRL NNPSRCHFGI VGSIYNLNDS RPFSLVDMMK PYNYLYDAIH DRLNKAIASN WGSILELDLS KVPKGWDVGK WMYYA RVNH IAVIDSFKEG TIGASTGKLA GALNNAGKGM IETNIGNYIQ QQINLLEFIK MEMADVAGIS KQREGQISQR ETVGGV ERA TLQSSHITEW LFTIHDDVKK RALECFLETA KVALKGRNKK FQYILSDTST RVMEIDGDEF AEADYGLVVD NSNGTQE LQ QKLDTLAQAA LQTQTLSFST ITKLYTSSSL AEKQRLIEKD EKQIRERQAQ AQKEQLEAQQ QIAAMQQQQK EAELLQKE E ANIRDNQTKI IIAQIQSEGG PDEEDGIMID DYSPEAKANL AEKIREFDEK LKLDKDKLKL DKKKAETDAS IKRQALRKK SSTTNK |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.3 µm Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.3 µm |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 51.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 157 |
|---|---|
| Output model |  PDB-7qoh: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)